Next Generation Sequencing: Unusual Cases of Spastic Paraplegic Presenting with Ataxia
Priyanka Vishwakarma, Sarita Agarwal, Deepika Delsa Dean and Kausik Mandal
DOI10.36648/2380-7245.21.7.40
Department of Medical Genetics, Sanjay Gandhi Postgraduate Institute of Medical Sciences (SGPGIMS), Lucknow 226014, India
- *Corresponding Author:
- Sarita Agarwal
Department of Medical Genetics
Sanjay Gandhi Postgraduate Institute of Medical Sciences (SGPGIMS)
Lucknow 226014, India
Tel: 91- 522-2494349
E-mail: saritasgpgi@gmail.com
Received Date: October 01, 2021; Accepted Date: October 15, 2021; Published Date: October 22, 2021
Citation: Vishwakarma P, Agarwal S, Dean DD, Mandal K (2021) Next Generation Sequencing: Unusual Cases of Spastic Paraplegic Presenting with Ataxia. J Rare Disord Diagn Ther Vol.7 No.10:40
Abstract
Hereditary Spastic Paraplegias (HSPs) are a group of heritable conditions resulting from neuro-degeneration of the cortico-spinal tracts. It can exist as “pure form” characterized by advanced spasticity, weakness of hind limbs, bladder dysfunctioning and mild somatosensory deficits, or as “complex form” where other neurological and systemic abnormalities are present.
The extensive clinical and genetic heterogeneity of spastic paraplegia and clinical overlap with various ataxias makes clinical diagnosis challanging. In this study, 70 unusuall ataxia cases were enrolled. Molecular diagnosis identified and excluded 18 positive cases of Spino Cerebellar Ataxia (SCA) subtypes and 1 positive case of Friedreich Ataxia (FRDA). Furthermore, from remaining uncharacterized 51 ataxia cases, we carried out Clinical Exome Sequencing (CES) in 10 individuals who depicted early age of onset with severe ataxia. CES identified 3 male cases with variants in SPG7, SPG11 and FA2H genes implicated in spastic paraplegia. Variants identified are: a heterozygous pathogenic variant c.2014G-A (p. Gly 672Arg) SPG7 gene (in case 1), a homozygous pathogenic variants c.869+1 G-T(5’Splice site), in SPG11 gene (case 2) and a heterozygous pathogenic c.1A-G (p.met1?) in FA2H gene (case 3). Thus CES facilitated in getting a definitive diagnosis in 3 out of 10 unusual HSP cases presenting with ataxia.
Keywords
Ataxia; Spastic paraplegia; Next generation sequencing; Sanger sequencing; Clinical exome sequencing
Introduction
Hereditary Ataxia (HA) is a collection of genetic anomalies categorized by slow progression of the movement incoordination and is often associated with reduced incoordination of hands, speech, and eye movements [1]. Also cerebellar degeneration is frequently observed in these disorders. Molecularly, many HA are caused by repeat expansion mutations as in case of Friedreich Ataxia (FRDA, OMIM 229300) and in different subtypes of Spino Cerebellar Ataxia (SCA) [1].
FRDA is an autosomal recessive neurodegenerative disorder characterized by progressive gait disturbances; limb ataxia associated with the weakness of lower limb and fore limb muscles and also absence of lower limb reflexes, dysarthria, and reduced vibratory senses and proprioception [2]. The commonest molecular abnormality, associated with FRDA is a Trinucleotide repeat (GAA) expansion in the intron 1 of FXN1 gene. Another type of HA is Spino Cerebellar Ataxia (SCA, OMIM 164400) which is a rare, genetically heterogeneous, neurodegenerative disorder with multiple subtypes. A majority of SCA subtypes are caused by dominant CAG expansion mutations (in coding and/ or noncoding region), while others are caused by truncating, missense and nonsense mutations [3]. HA are also caused due to other single gene disorders such as Episodic Ataxias (EA) and Ataxia Talengiectasia (AT).
Hereditary Spastic Paraplegias (HSPs) are a group of heritable conditions resulting from neuro-degeneration of the corticospinal tracts and is referred as “pure form” when the signs and symptoms are limited to advanced spasticity, weakness of the hind limbs, dysfunctioning of the bladder and somatosensory deficits in mild form. While in “complex form”, neurologic impairment and other system involvement are also present apart from those present in pure form. Till date, seventy different HSP loci, and about sixty causative genes have been identified [4]. The mode of inheritance of HSP ranges from autosomal dominant (AD)/autosomal recessive (AR))/X-linked to mitochondrial [4].
Patients who display complex form of HSP may present with cerebellar ataxia along with spastic paraplegia phenotype and thus often represents diagnostic challenge, as the differential diagnosis is very broad. Therefore implementing Clinical Exome Sequencing (CES) is highly justified in order to reach to a diagnostic conclusion in such cases [5]. In this study, we have recruited clinical ataxia suspected cases for CES leading to identification and molecular classification of three unrelated, unusual spastic paraplegia cases among them.
Methods
Sample size
Initially we have recruited 70 clinical suspects of HA (age range 16 to 70 years) of Indian origin. SCA subtypes that are common in India viz. SCA subtypes 1, 2, 3, 6 and 7 and FRDA are ruled out by molecular testing. After confirming diagnosis of FRDA and SCA positive cases, we selected 10 cases from the remaining uncharacterized cases and subjected them to CES analysis (covering 6,000 genes related to neurological disorders).
Inclusion and exclusion criteria for CES testing are as follows:
Inclusion Criteria:
• Patients presenting severe ataxia related features.
• Patients presenting an early age of onset of ataxic like features.
Exclusion Criteria:
Patients positive for Spino Cerebellar Ataxia (SCA) and Freidreich Ataxia (FRDA).
Sample collection and DNA isolation:
Two millilitres of peripheral blood were collected in EDTA vial. Genomic DNA was isolated by Qiagen DNA extraction kit according to the manufacturer's instructions (Qiagen, Valencia, CA). The quality and quantity of DNA were assured by agarose gel electrophoresis and nanodrop 2000 (Thermo Scientific, USA), respectively.
Molecular analysis
PCR based analysis: We excluded triplet expansion mutation in the genes causing SCA type 1, 2, 3, 6 and 7 (ATXN1, ATXN2, ATXN3, CACNA1A, ATXN7) utilizing Multiplex PCR (M-PCR) and FRDA (FXN1) by using TP-PCR as per the method of Dorschnerm, et al. [6] and Bhowmik et al. [7], respectively.
Clinical Exome Sequencing: Targeted capture and sequencing of the coding region’s protein of the genome/genes were performed. Genomic DNA was used to achieve the targeted gene capture by expending a custom capture kit. The libraries were sequenced to mean >80-100 X coverage on Illumina sequencing platform. The sequences obtained were aligned to human reference genome (GRCh37/hg19). Clinically relevant mutations were annotated using published variants in literature and a set of diseases databases - ClinVar, OMIM (updated on 21st November 2018), GWAS, HGMD (v2018.3) and SwissVar [8-12]. Common variants were filtered based on allele frequency in 1000 Genome Phase 3, ExAC (v1.0), gnomAD (v2.1), EVS, dbSNP (v151) in one thousand Japanese Genome and in our internal Indian population databases [13-17]. Non-synonymous variants effect is calculated using multiple algorithms such as PolyPhen-2, SIFT (Scale-invariant Feature Transform, MutationTaster2 and LRT. Only non-synonymous and splice site variants found in the clinical exome panel consisting of 8332 genes were used for clinical interpretation. Silent variations that do not result in any change in amino acid in the coding region were not reported.
Sanger Sequencing: Sanger sequencing was performed for confirmation and validation of the variants obtained by CES analysis.
Results
Among the 10 patients enrolled for CES, we found mutations in 3 out of 10 patients, in 3 different genes implicated in spastic paraplegia. Following the identification of mutations in spastic paraplegia genes, we re-evaluated these cases to see if their phenotypes fit into the underlying disorder. The details are described below.
Case 1
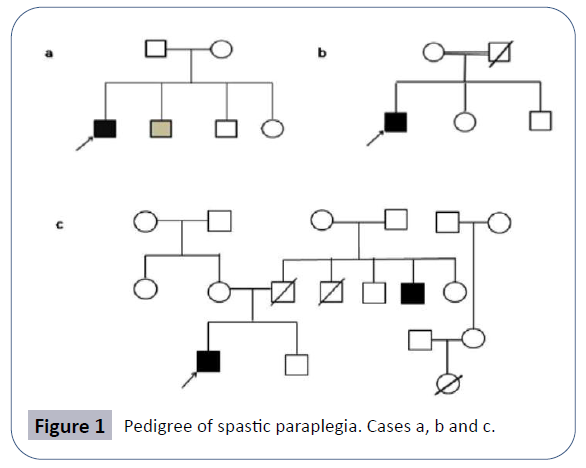
AK was a 24-year-old male, born to non-consanguineous parents of Indian origin who hailed from Uttar Pradesh, after an abnormal pregnancy of forty weeks with normal birth dimensions. Proband’s height was 160.02 centimeters with 65 kg weight (Case 1). Both parents were healthy (Figure 1). But the younger brother presented mild symptoms with walking problem due to lower limb weakness. The proband’s sister complained mild knee pain. Family pedigree of patient is shown in (Figure 1b).
Proband reported walking difficulty, whole body balancing difficulty and spasticity in the lower limbs at 16 years of age as his first symptoms and later the symptoms get aggravated like the pain in the waist region, problems related to sitting and walking. The patient also noticed gastric problem. MRI suggested the classic pattern of spastic paraplegia with the occurrence of bilaterally hyper intense signals in the basal ganglia, thalami and of the Periventricular white matter. MRI also showed the diffuse disc bulge with broad based postero-central prostrusion at LS-S1 level producing mild to moderate extradural compression over ventral aspect of thecal sac and causing mild narrowing of lateral recesses and neural foramina of both side.
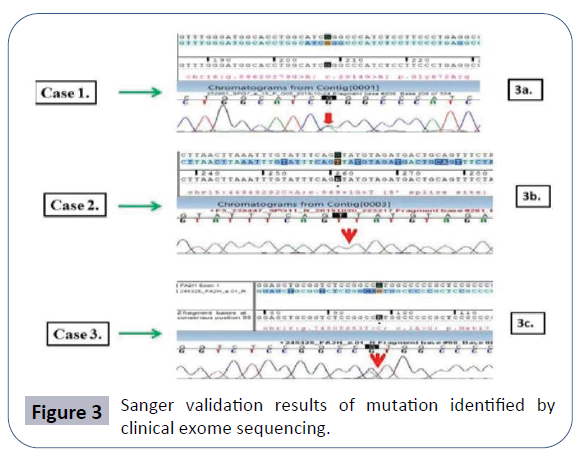
Diffuse disc bulge is noted at L2-3, L3-4 and L4-5 levels producing mild extradural compression over ventral aspect of thecal sac and causing mild narrowing of bilateral neural foramina. Early MRI finding suggested a spondo-discal degenerative change in lumber spine as described above. In this patient, the Clinical exome sequencing identified the heterozygous missense variation in the exon number 15 of SPG7 gene (chr16:g.2014G>A) that results in substitution of the amino acid glycine to arginine at codon number 672 (p.Gly672Arg; ENST00000268704.2) (Figure 3a).
Case 2
MR was a 24-year-old male born to consanguineous parents of the Indian origin hailing from Uttar Pradesh, after a normal pregnancy with normal birth measurements. Proband’s height is 162.5 centimeters and with 70 kg weighing (Case 2). Both parents and siblings (one brother and one sister) are healthy. Problem in writing, walking and whole body movement and stiffness in lower limbs appeared at 15 years of age as the first symptoms of the disease after this the symptoms progressed gradually like the pain in waist region and problem in sitting and walking. Family pedigree is shown in (Figure 1b).
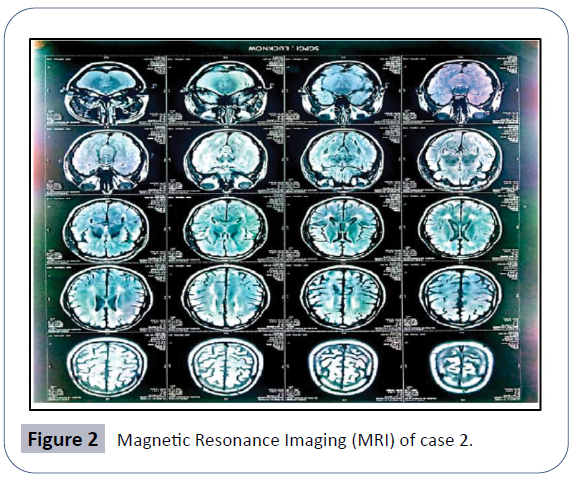
The MRI finding observed T2 flair hypersensitivity and degeneration in corona radiate and periventricular white matter, the typical pattern of spastic paraplegia (Figure 2). No metabolic problem so far detected in patient.
In this patient, the clinical exome sequencing identified a homozygous 5' splice site variation in intron 4 of the SPG11 gene (chr15:g.44949292C>A; Depth: 18 X) that affects the invariant GT donor splice site of exon 4 (c.869+1G>T; ENST00000261866.7) (Figure 3b and Case 2).
Case 3
PS was 19 year-old male born to non- consanguineous parents of Indian origin and hailing from Bihar. The pregnancy was normal with normal birth measurements. Proband’s height was 160.02 centimeters and with 65 kg weight (Figure 4 and Case 3), both parents were healthy.
The clinical features presented by proband were scanning speech, behavioral changes since last 6 years, irritation, anger, irrelevant talking, sense of isolation, aggressive behavior, gait ataxia, and falling suddenly while walking and weakness in lower limbs. Problem in walking and whole body balancing was appeared at 10 years of age as the first symptoms of the disease after this the symptoms got worsened (Figure 1c). The MRI report indicated (not shown here) the distinctive patterns of the spastic paraplegia with the occurrence of thin corpus callosum.
In this patient CES detected a start-loss variation in exon 1 of the FA2H gene (chr16:g.c.1A>G; Depth: 3x) that altered the ATG start codon and consequently affected its translation (p.Met1?; ENST00000219368.3) (Figure 3c).
For all the three variants identified by CES validation of the result was carried out by Sanger sequencing and is shown in (Figure 3a- 3c) respectively.
Discussion
The brain is the most important part of the Central Nervous System (CNS), and about 84% of the genes are expressed in the brain region [18]. A little alteration in expression of the genes in brain could lead to severe concern and is implicated in numbers of neurological problems inclusive of HA and HSPs. HSP or cortico-spinal motor neuron disease is a heterogeneous group of degenerative disorders characterized by progressive weakness and spasticity of the lower limbs, combined with additional neurological features.
The extensive clinical and genetic heterogeneity with over 80 potential disease-associated genes and frequent overlap with other clinical conditions affecting the motor system makes molecular diagnosis in HSP cumbersome and time consuming. Thus in many HSP cases the diagnosis remained unconfirmed in spite of a huge number of independent molecular diagnostic tests series after the clinical diagnosis. Population based studies have depicted that some HSP implicating mutations are quite common in several populations, while some are very rare ones in other populations [19], thus requiring panel based testing. Confirmatory molecular diagnosis is very important for the ultimate diagnosis in the affected individuals, for giving the surety and avoiding the needless molecular diagnostic
Clinical exome sequencing approaches are increasingly being used for genetic diagnostics in routine clinical settings, and different published papers report successful use of this technology in HSP [20-23], with positive yields ranging from 20% in adult cases to 52.5% in child cohorts [23-27]. In this study, we have been able to identify genetic mutations related to HSP in 3/10 (~44 %) unexplained ataxia patients by CES. The first patient (Case 1) confirmed for HSP had a heterozygous missense variation in the exon number 15 of SPG7 gene (chr16:g.89620279G >A) that resulted in substitution of the amino acid glycine at codon number 672 to arginine (p.gly 672Arg; ENST00000268704.2). The p.Gly 672 Arg variant has not been reported in the 1000 genomes and has a minor allelic frequency of 0.0008% in ExAC Database.
The variation identified in Case 1 had previously been reported, but in compound heterozygous state along with other missence mutation (c.1529C >T) in a patient affected with adult onset upper motor neuron syndrome in Dutch patients [28]. Spastic paraplegia-7 is caused by mutation in SPG7 gene. The SPG7 gene delivers the instructions for making a paraplegin protein, a member of the AAA protein family [29]. AAA protein family plays an important role in the several cellular activities, such as regulation of the cellular components and the proteins.
These proteins is located within the inner membrane of the mitochondria., As paraplegin is highly expressed in Purkinje neurons which are involved in movement coordination, thus it explains the manifestation of ataxic feature in Case 1 with SPG 7 mutation [30]. Mutations in SPG7 are responsible for 1.5–4.5% of autosomal recessive HSP cases with both, pure and complicated phenotypes [31]. Thus it has been suggested that SPG7 should be analyzed when autosomal recessive form of adult onset complex HSP is suspected [32]. Warnecke T et al. [33] had previously reported a missense mutation c.2075G>C in exon 15 of the SPG7 gene in the homozygous state. However Dominant effect of SPG7 has also been identified in some significant studies and challenged this concept. In our study we have identified a heterozygous mutation in exon 15 of SPG7 gene upon querying 6000 genes movement disorder related CES panel, hence suggesting a dominant inheritance with variable penetrance pattern for this gene [34].
In second patient (Case 2), A homozygous 5' splice site variation in intron 4 of the SPG11 gene (chr15:g.44949292C >A; Depth: 18 X) that affects the invariant GT donor splice site of exon 4 (c.869+1G >T; ENST00000261866.7) was detected. The observed variation had been previously reported (as 869+1G >A) in patients affected with spastic paraplegia [35]. For this variant the in silico prediction is the damaging causing by the MutationTaster2 software. The reference base is also conserved across mammals in the databases. Gene SPG11 codes 8-kb mRNA consisting of 40 exons and the protein expresses in adult cerebellum, cerebral cortex, hippocampus and the pineal gland region of the brain [36]. Protein encoded by SPG11 gene is the Spatascin consists of 2443 amino acids with indefinite functions. It has been suggested that Spatacsin play a vital biological role due to its high conservation among species [37]. Kara et al., 2016 reported the mutations affecting the SPG11 gene as the main reason of the autosomal recessive HSP responsible for approximately twenty five % of the cases [38].
The third patient (Case 3), was identified to have a heterozygous start-loss variation in exon 1 of the FA2H gene (chr16:g.74808653T>C; Depth: 3 X) that alters the ATG start codon and consequently affects its translation (p.Met1?; ENST00000219368.3).The p.Met1? Variant has not been reported in the 1000 genomes, ExAC and databases. This variant is predicted to be damage causing by the software SIFT and the MutationTaster2. The reference codon is conserved across mammals. FA2H gene is located on chromosome 16q23. FA2H gene encodes for the endoplasmic reticulum enzyme fatty acid 2-hydroxylase (FA2H) that plays significant role in the formation of 2-hydroxy glycolsphingolipids (major myelin component). FA2H homozygous mutations is implicated in neurodegeneration with iron accumulation in brain (fatty acid hydroxylase-associated neurodegeneration, FAHN), HSP type SPG35 and leukodystrophy with spasticity and dystonia [39,40], but not much about the functional impact of heterozygous FA2H mutations is known. Recently, it has been reported that rare deleterious heterozygous mutations of FA2H might constitute risk factors for Autism Spectrum Disorder (ASD) [41]. Other studies suggest FA2H heterozygous mutations as minor risk factors for ASD.
Clinical investigation in Case 3 depicted behavioral impairment and social aversion apart from ataxia and movement impairment. We may speculate that some other additional mutations in genes related to autism or intellectual disability might be present. Alternatively, as the effect of mutation on FA2H is not known but predicted deleterious with various in silico tools, thus it may be directly involved in ASD phenotype as well. Our study for the first time showed movement related phenotype (presence of ataxia symptoms) along with mild ASD like features related to heterozygous FA2H mutation.
The extensive clinical and genetic heterogeneity of spastic paraplegia and the clinical overlap between various ataxia cases suggests that it is difficult to arrive at a rapid and precise diagnosis of these conditions and requires definitive diagnosis. The Past studies for classification of HSP cases have been done by accomplishment of the CES or the WES (whole-exome sequencing) [42-44]. CES results generally offer precious information which can be used for the clinical resolution making.
In our study we studied 3 patients with a pure form of unexplained ataxia by CES and found 3 patients positive for mutations in genes implicated in HSP (SPG7, FA2H and SPG11). Among them 2 were found to have heterozygous mutation in SP7 and FA2H genes, whose recessive mutations were known to be linked with spastic paraplegia respectively. Third patient had a homozygous variant in SPG11 gene and is in concordance with previous reports. As these patients were initially clinically characterized as ataxia suspects thus it is evident that the clinical continuum [45-46] of the hereditary spastic paraplegia‘s variants are extended to the pure cerebellar ataxias also.
Conclusion
In the era of medical genomics and precision medicine, which has brought the first randomized clinical trials in HSP and a deeper approach in modern neuro rehabilitation, high levels of genetic heterogeneity should no longer prolong the time to diagnosis and preclude access to new treatment and care opportunities. We consider that the Clinical Exome Sequencing will be a worthwhile tool for the diagnosis of known Mendelian genetic diseases such as HSP and HA.
References
- Bird TD, Adam MP, Ardinger HH, Pagon RA (2020) Hereditary ataxia overview. Genereviews® [Internet]. Seattle (WA): University of Washington, Seattle.
- Parkinson MH, Boesch S, Nachbauer W, Mariotti C, Giunti P (2013) Clinical features of Friedreich's ataxia: Classical and atypical phenotypes 1:103-17.
- Jacob J (2002) Hansen hereditary spastic paraplegia SPG13 Is Associated with a mutation in the gene encoding the mitochondrial chaperonin Hsp60. Am J Hum Genet 70:1328-1332.
- Hedera P, Adam MP, Ardinger HH, Pagon RA (2019) Hereditary spastic paraplegia overview. University of Washington, Seattle.
- Van De Warrenburg B, Schouten M, De Bot S, Vermeer S, Meijer R, et al. (2016) Clinical exome sequencing for cerebellar ataxia and spastic paraplegia uncovers novel gene disease associations and unanticipated rare disorders. Eur J Hum Genet 24:1460-1466.
- Dorschner MO, Barden D, Stephens K (2002) Diagnosis of five spinocerebellar ataxia disorders by multiplex amplification and capillary electrophoresis. J Mol Diagn 4:108‐113.
- Das Bhowmik A, Rangaswamaiah S, Srinivas G, Dalal AB (2015) Molecular genetic analysis of trinucleotide repeat disorders (TRDS) in Indian population and application of repeat primed PCR. Eur J Med Genet 58:160-167.
- Li H (2010) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589-95.
- Mclaren W (2010) Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics.P.2069-70.
- Zerbino DR (2018) Ensembl. Nucleic Acids Res 46:D754-D761
- Plagnol V (2012) A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics 28:2747-2754.
- Landrum MJ (2015) Clinvar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res 44:D862-8.
- Mckusick VA (2007) Mendelian Inheritance in Man and its online version OMIM. Am J Hum Genet 80: 588‐604.
- Welter D (2014) The NHGRI GWAS catalog: A curated resource of SNP-trait associations. Nucleic Acids Res 42: D1001-1006.
- Stenson PD (2017) The human gene mutation database: Towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next generation sequencing studies. Human Genet 136:665-677
- Mottaz A (2010) Easy retrieval of single amino-acid polymorphisms and phenotype information using SWISSVAR. Bioinformatics 26:851-852.
- (2015) 1000 genomes project consortium. A global reference for human genetic variation. Nature. 526: 68-74.
- Hawrylycz MJ, Lein ES, Guillozet Bongaarts AL, Shen EH, Ng L, et.al (2012) An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489:391-399.
- Erichsen AK, Koht J, Stray Pedersen A, Abdelnoor M, Tallaksen CM (2009) Prevalence of hereditary ataxia and spastic paraplegia in southeast Norway: A population-based study. Brain 132: 1577-1588.
- Balicza P, Grosz Z, Gonzalez MA, Bencsik R, Pentelenyi K, et al. (2016) Genetic background of the hereditary spastic paraplegia phenotypes in Hungary an analysis of 58 probands. J Neurol Sci 364:116-21.
- Bettencourt C, López-Sendón JL, García-Caldentey J, Rizzu P, Bakker IM, et al. (2014) Exome sequencing is a useful diagnostic tool for complicated forms of hereditary spastic paraplegia. Clin Genet 85:154-8.
- Kumar KR, Blair NF, Vandebona H, Liang C, Ng K, et al. (2013) Targeted next generation sequencing in SPAST-negative hereditary spastic paraplegia. J Neurol 260:2516-22.
- Lynch DS, Koutsis G, Tucci A, Panas M, Baklou M, et al. (2016) Hereditary spastic paraplegia in Greece: Characterisation of a previously unexplored population using next-generation sequencing. Eur J Hum Genet 24:857-63.
- Kara E, Tucci A, Manzoni C, Lynch DS, Elpidorou M, et al. (2016) Genetic and phenotypic characterization of complex hereditary spastic paraplegia. Brain 139:1904-18.
- Schüle R, Wiethoff S, Martus P, Karle KN, Otto S, et al. (2016) Hereditary spastic paraplegia: Clinicogenetic lessons from 608 patients. Ann Neurol 79:646-58.
- Burguez D, Polese-Bonatto M, Scudeiro LAJ, Björkhem I, Schöls L, et al. (2017) Clinical and molecular characterization of hereditary spastic paraplegias: A next-generation sequencing panel approach. J Neurol Sci 383:18-25.
- Iqbal Z, Rydning SL, Wedding IM, Koht J, Pihlstrøm L, et al. (2017) Targeted high throughput sequencing in hereditary ataxia and spastic paraplegia. Plos ONE 12: e0174667 10.
- Brugman F (2008) Paraplegin mutations in sporadic adult-onset upper motor neuron syndromes. Neurology 1:526-632.
- Arnoldi A, Tonelli A, Crippa F, Villani G, Pacelli C, et al. (2008) A clinical, genetic and biochemical characterization of SPG7 mutations in a large cohort of patients with hereditary spastic paraplegia. Hum Mutat 29:522.
- Sacco T, Boda E, Hoxha E, Pizzo R, Cagnoli C, et al. (2010) Mouse brain expression patterns of Spg7, Afg3l1, and Afg3l2 transcripts, encoding for the mitochondrial m-AAA protease. BMC Neurosci P.55.
- Bhattacharjee S, Beauchamp N, Murray BE, Lynch T (2017) Case series of autosomal recessive hereditary spastic paraparesis with novel mutation in SPG 7 gene. Neurosciences (Riyadh) 22:303-307.
- Hedera P, Adam MP, Ardinger HH, Pagon RA (2020) Hereditary spastic paraplegia overview. Genereviews® [Internet]. Seattle (WA): University of Washington, Seattle.
- Warnecke T, Duning T, Schwan A, Lohmann H, Epplen JT, et al. (2007) A novel form of autosomal recessive hereditary spastic paraplegia caused by a new SPG7 mutation. Neurology 69: 368-375.
- Tesson C, Koht J Stevanin G (2015) Delving into the complexity of hereditary spastic paraplegias: How unexpected phenotypes and inheritance modes are revolutionizing their nosology. Hum Genet 134:511-538.
- Lek M (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536:285-91.
- Stevanin G, Santorelli FM, Azzedine H (2007) Mutations in SPG11, encoding spatacsin, are a major cause of spastic paraplegia with thin corpus callosum. Nat Genet 39 : 366-372.
- Orlacchio A, Babalini C, Borreca A, Patrono C, Massa R, et.al (2010) SPATACSIN mutations cause autosomal recessive juvenile amyotrophic lateral sclerosis. Brain 133:591-598.
- Kara E, Tucci A, Manzoni C (2016) Genetic and phenotypic characterization of complex hereditary spastic paraplegia. Brain 139:1904-1918.
- Edvardson S, Hama H, Shaag A, Gomori JM, Berger I, et.al (2008) Mutations in the fatty acid 2-hydroxylase gene are associated with leukodystrophy with spastic paraparesis and dystonia. Am J Hum Gene 83:643-8.
- Kruer MC, Paisán Ruiz C, Boddaert N, Yoon MY, Hama H (2010) Defective FA2H leads to a novel form of neurodegeneration with brain iron accumulation (NBIA). Ann Neurol 68:611-8.
- Scheid I, Maruani A, Huguet G, Leblond CS, Nygren G, et.al (2013) Heterozygous FA2H mutations in autism spectrum disorders. BMC Med Genet 14: 124.
- Elert Dobkowska E, Stepniak I, Krysa W (2019) Next generation sequencing study reveals the broader variant spectrum of hereditary spastic paraplegia and related phenotypes. Neurogenetics 20: 27.
- Iqbal Z, Rydning SL, Wedding IM, Koht J, Pihlstrøm L (2017) Targeted high throughput sequencing in hereditary ataxia and spastic paraplegia. Plos ONE 12: e0174667.
- D’Amore A, Tessa A, Casali A, Teresa Dotti M, Alessandro Filla (2018) Next generation molecular diagnosis of hereditary spastic paraplegias: An Italian Cross-Sectional Study. Front Neurol 9:981
- Marelli C, Lamari F, Rainteau D, Lafourcade A, Banneau G, et al. (2018) Plasma oxysterols: Biomarkers for diagnosis and treatment in spastic paraplegia type 5. Brain 141:72-84.
- Serrao M, Chini G, Bergantino M, Sarnari D, Casali C, et al. (2018) Identification of specific gait patterns in patients with cerebellar ataxia, spastic paraplegia, and Parkinson's disease: A non-hierarchical cluster analysis. Hum Mov Sci 57:267-79.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences