Clinical and Molecular Spectrum of Sarcoglycanopathy Related Genes in Iranian Families
Mohammad Amin Khodadadegan1, Soheila Abedini2, Mohammad Doosti2, Paria Najarzadeh Torbati2, Mojtaba Safi2, Hadis Malek2, Samaneh Maskani2, Masoumeh Ramahi2, Sogand Malekzadeh3, Mehran Beiraghi Toosia4, Reza Boostani5, Najmeh Ahangari6, Ehsan Ghayoor Karimiani2,6,7* and Yalda Jamshidi7
1Department of Student Research Committee, Mashhad University of Medical Sciences, Mashhad, Iran
2Department of Molecular Genetics, Next Generation Genetic Polyclinic, Mashhad, Iran
3Department of Biology, Mashhad Branch, Islamic Azad University, Iran
4Department of Pediatric Neurology, Ghaem Hospital, Mashhad University of Medical Sciences, Mashhad, Iran
5Department of Neurology, Mashhad University of Medical Sciences, Mashhad, Iran
6Department of Innovative Medical Science, Mashhad Branch, Islamic Azad University, Mashhad, Iran
7Department of Molecular and Clinical Sciences, St. George's, University of London, Cranmer Terrace, Iran
- *Corresponding Author:
- Ehsan Ghayoor Karimiani
Department of Molecular, and Clinical Science, St. George's University of London, Cranmer Terrace, London SW17 0RE, UK
Tel: 989153102327
E-mail: ngc.article@gmail.com
Received Date:March 14, 2022, Manuscript No. IPRDDT-22-12387; Editor assigned date: March 17, 2022, PreQC No. IPRDDT-22-12387 (PQ); Reviewed date: March 31, 2022, QC No. IPRDDT-22-12387; Revised date: May 11, 2022, Manuscript No. IPRDDT-22-12387 (R); Published date: May 18, 2022, DOI: 10.36648/2380-7245/8.5.56
Citation: Karimiani EG, Khodadadegan MA, Abedini S, Doosti M, Torbati PN, et al. (2022) Clinical and Molecular Spectrum of Sarcoglycanopathy Related Genes in Iranian Families. J Rare Disord Diagn Ther Vol:8 No:5
Abstract
Sarcoglycanopathies comprise four subtypes of autosomal recessive limb-girdle muscular dystrophy (LGMD2C, LGMD2D, LGMD2E, and LGMD2F) caused, respectively, by mutations in the SGCG, SGCA, SGCB, and SGCD genes. Knowledge about the clinical and genetic features of sarcoglycanopathies in Iranian patients is limited. This study aimed to investigate the clinical manifestations and gene mutations in Iranian patients with sarcoglycanopathies and identify possible correlations.
Studies on SGCs were performed on ten unrelated Iranian families referred to a genetic clinic for genetic diagnosis due to the initial diagnosis of muscular dystrophy. Based on clinical phenotype and genetic findings were observed, respectively: The ten unlinked LGMDs families originated from 9 different cities of Iran. Three patients were diagnosed with LGMD2D, four with LGMD2E, and three with LGMD2C. No patient was detected with LGMD2F. Ten mutations were detected in SGCA (n=3), SGCB (n=4), and SGCG (n=3); no SGCD mutation was found in these families. The finding will be beneficial for screening and genetic counseling of SGCs patients in Iran.
The present study demonstrates the clinical and molecular spectrum of Sarcoglycanopathy related-genes in the study families. Our finding provided additional insights into genotype and phenotype correlations in the Iranian population.
Keywords
Sarcoglycanopathy; Limb-girdle muscular dystrophy; WES
Introduction
The Limb Girdle Muscular Dystrophy (LGMD) includes many Mendelian disorders categorized by progressive degeneration of proximal limb muscles. The shoulders and hip region are the earliest affected muscles. Sarcoglycanopathies (SGCs) are subgroups of autosomal recessive LGMD that occur due to debilitating mutations in SGCA, SGCB, SGCG, and SGCD genes encoding the four types of the voluntary muscle sarcoglycan complex, alpha, beta, gamma, and delta-sarcoglycan sarcoglycan (transmembrane glycoproteins), which lead to LGMD2D, 2E, 2C, 2F, respectively [2]. These sarcoglycans form the stability of the dystrophin-dystroglycan complex and plasma membrane cytoskeleton [3]. The prevalence of sarcoglycanopathies differs between populations based on race and geographic region. LGMD2E and LGMD2D have the most and lowest frequency of sarcoglycanopathies in the Iranian, respectively [4]; while, LGMD2D is relatively prevalent in the US and Europe [5-6]. LGMD2C is the most common type of LGMD among Algerian and Indian populations [7-8]. Overall, 16 mutated genes have been reported to be involved with various forms of LGMD [6].
SGCs manifest different clinical features ranging from mild to severe [9]. The onset age of symptoms usually occurs between five and fifteen years old [10]. The primary symptoms of the diseases include weakness of the shoulder and pelvic girdle muscles; other manifestations may be revealed by calf hypertrophy, scapular winging, lumbar hyperlordosis, and a higher plasma level of creatine kinase [10]. In the last stages of the disease, SGCSs often involve respiratory and cardiac systems [11]. SGCs resemble the Intermediate type of Duchenne and Becker Muscular Dystrophy [11-13]. Considering difficult differentiation among subtypes of SGCs depending on clinical manifestations, thus a combination of immunoblot, immunohistochemical analyses, and subsequent DNA sequencing have been proposed for accurately diagnosing the underlying pathology [14].
However, there is no definitive cure to attenuate muscle weakness and reverse mobility function of patients with limb-girdle muscular dystrophy; Supportive treatment should be adapted to each patient and each subtype of LGMD [15]. Accurate diagnosis of SGCs is a key factor in genetic counseling, preimplantation genetic diagnosis, and prospective therapies.
Lastly, studies on the SGCs have been conducted among Iranian patients by genetics analysis [16-19]. To the best of our knowledge, previously published studies about the clinical features and genotype of SGCs are limited in Iranian patients. We aimed to detail the clinical manifestations, pedigrees, genotype, and gene mutations in ten families of the Iranian population with sarcoglycanopathies.
Materials and methods
Patients and Samples
Probands were selected from ten unrelated families referred to the genetic clinic for genetic counseling due to a primary diagnosis of muscular dystrophy. Written informed consent was obtained from all the study participants or their parents. 5 ml of peripheral blood with EDTA were obtained from patients and other available family members. DNA was extracted using standards salting out protocol. The proband' DNA samples were sent to Macrogen Company (Korea) for the Whole-Exome Sequencing (WES) 100X.
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Genetic testing and bio-informatics analysis
After data analysis, the Polymerase Chain Reaction (PCR) technique was performed using specific primers to confirm the candidate genetic variants, followed by Sanger sequencing. We interpreted and classified sequence variants through the Human Gene Mutation Database (HGMD) and ClinVar. In the case of a novel variant, it was classified as pathogenic, likely pathogenic, uncertain significance, likely benign, or benign according to the American College of Medical Genetics and Genomics (ACMG, 2015) guideline.
Results
Clinical phenotype
Table 1 summarizes the clinical features of patients with sarcoglycanopathies.
| ID | LGMD subtype | Age/sex | Onset age (years) | Disease duration years | Symptom(s) at onset | Current added symptoms | complications | AST | LDH | CK (IU/L) |
|---|---|---|---|---|---|---|---|---|---|---|
| F1 | LGMD2D | 10/F | Difficulties in standing and walking | Difficulties in standing and walking | NA | NA | NA | 1200 | ||
| F2 | LGMD2D | 13/M | 6 | 7 | Difficulties in climbing stairs post walking muscle pain upper limb weakness | Difficulties in standing and walking | Lordosis | - | - | 11000 |
| F3 | LGMD2E | 12/F | 4 | 8 | Difficulties in running, standing, and walking | Unable to stand(wheelchair) | Lordosis | - | - | 1550 |
| F4 | LGMD2C | 17/F (F4-P1) | 9 | 8 | Early fatigue calf stiffness difficulties in standing | Unable to stand(wheelchair) upper limb weakness | Lordosis | 1110 | - | 16390 |
| 9/F (F4-P2) | 8 | 1 | Calf stiffness | Difficulties in standing and running upper limb weakness | Lordosis | - | 780 | 23040 | ||
| F5 | LGMD2C | 17/M | 6 | 11 | Running difficulty | Difficulties in standing upper limb weakness | Cardiovascular Lordosis Scoliosis | - | 99.8 | 4635 |
| F6 | LGMD2E | 6/F | 6 | Proximal lower limb weakness, early fatigue, and calf hypertrophy | Thigh hypertrophy | - | 437 | 5200 | 23665 | |
| F7 | LGMD2E | 7.7/M (F7-P1) | 0.6 | 7.1 | Difficulties in swallowing chronic diarrhea | Difficulties in standing, running and climbing upstairs Achilles tendinitis | Lordosis Respiratory disorder | 323 | - | 23880 |
| 3.7/F (F7-P2) | 2.7 | 1 | Asymptomatic hyper CKemia | Difficulties in climbing stairs, itching | - | 226 | - | 14014 | ||
| F8 | LGMD2C | 7/6/M | 6.3 | 1.3 | Myoglobinuria | Difficulties in standing post-walking muscle pain flank pain | Lordosis scoliosis respiratory disorder | - | - | 21150 |
| F9 | LGMD2E | 46/F | 18 | 28 | Early fatigue frequent fall proximal lower limb weakness foot pain lower back pain | Falling in walking lower limb atrophy | Lordosis | 496 | - | 4342 |
| F10 | LGMD2D | 23/F (F10-P1) | 2 | 21 | Difficulties in running and standing. Falling upper limb weakness | Lower limb atrophy | Lordosis | - | - | 1500 |
| 2/M (F10-P2) | 0.2 | 1.8 | Drug-resistant epilepsy difficulties in holding head up | Difficulties in walking | - | - | - | 1300 |
Table 1: Clinical characteristics of study patients.
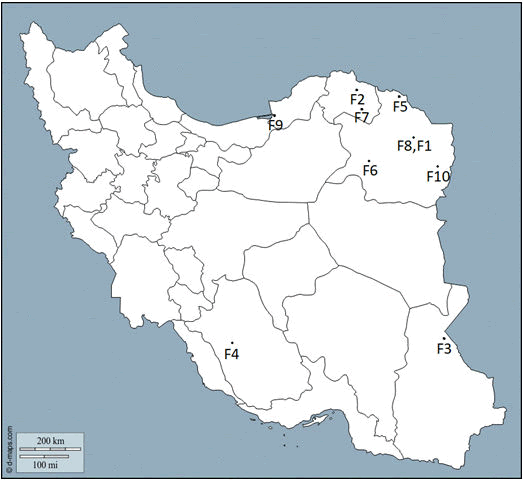
The ten unlinked LGMDs families originated from nine different cities of Iran, of which five were from nearby towns in a large province (Razavi Khorasan) in the northeast of Iran (Figure 1).
Families 1 and 8 are from the same small community/village and presumably related. Each patient's city of residence has been shown with a number. (1) Mashhad, (2) Bojnurd, (3) Zabol, (4) Shiraz, (5) Dargaz, (6) Kashmar, (7) Esfarayen, (8) Mashhad, (9) Bandar Torkaman, (10) Torbat-eJam.
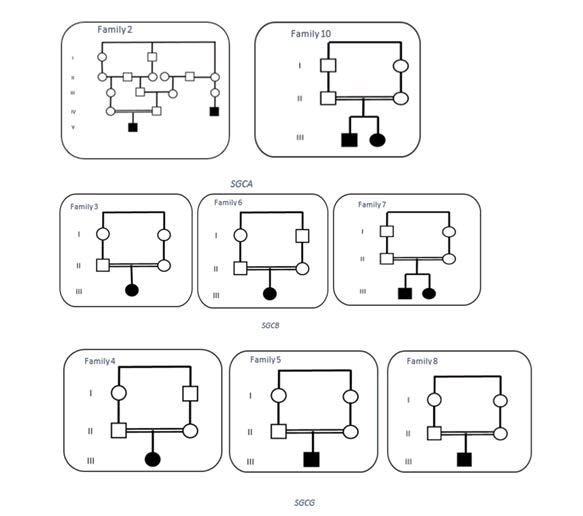
Three patients were diagnosed with LGMD2D, four with LGMD2E, and three with LGMD2C. No patient was detected with LGMD2F. Three families reported a positive family history in the other offspring, which is confirmed by paraclinical evaluations. Thirteen affected subjects from ten families were assessed clinically in this study. There were five males (38%) and eight females (61%). The consanguinity rate was 80%; in seven families, the parentswerefirstcousins(Figure 2).
In three patients (33%), the symptoms at the onset were associated with difficulties in running and calf stiffness; in two patients (20%), the sign at onset was early fatigue. The clinical presentation of the affected individuals was varied, rangingfromasymptomatic hyperkalemia to non-ambulatoryparaparesis. Difficulties in standing, walking, and climbing upstairs were the most common complaints among patients. Otherclinicalsymptoms not related to motor included chronic diarrhea, Drug-resistant epilepsy, and myoglobinuria. Lordosis was observed as a common complication in almost all patients. Likewise, one patient with LGMD2C suffereda cardiac abnormality (F5), and two patients with LGMD2E and LGMD2C had respiratory complications (F7-P1 and F8 respectively). CK concentrations were elevated in all patients. One patient was diagnosed with sarcoglycanopathy after an incidental finding of hyper CKemia (F7-P2). Two patients were no longer capable of ambulating independently (F3 and F4-P1) (Figure 3).
Genetic findings
According to Table 2 totally, ten mutations were detected in SGCA (n=3), SGCB (n=4), and SGCG (n=3), eight of them have already been reported pathogenic in genomic databases such as ClinVar and the remaining two variants were novel. No SGCD mutation was found in these families. All patients had a complete molecular diagnosis and were found to have only one mutation in SGCA, SGCB, or SGCG. The pathogenic mutations included six missense, two deletion, one-stop codon, and one splicing mutations. Except for the heterozygote mutations found in patient nine, other mutations were the homozygous state in different patients.
| Family | Gene | c.DNA position | Exon | Effect on protien | Type of variants | Zygosity | Parental | Variants Pathogenicity |
|---|---|---|---|---|---|---|---|---|
| F1 | SGCA | c.100C>T | Exon 2 | p. Arg34Cys | missense | Hom | Proband | Pathogenic |
| F2 | SGCA | c.113A>G | Exon 2/CDS2 | p. His38Arg | missense | Hom | Proband | VUS |
| F3 | SGCB | Exon 2 deletion | Exon 2 | - | deletion | Hom | Proband | Pathogenic |
| F4 | SGCG | Exon 3 deletion | Exon 3/CDS2 | - | deletion | Hom | Proband | Pathogenic |
| F5 | SGCG | c.704T>C | Exon 8/CDS7 | p. Leu235Pro | missense | Hom | Proband | Likely pathogenic |
| F6 | SGCB | c.544A>C | Exon4 | p. Thr182Pro | missense | Hom | Proband | VUS |
| F7 | SGCB | c.753+1G>A | Exon 5 | N/A | splicing | Hom | Proband | Likely pathogenic |
| F8 | SGCG | c.31G>T | Exon 2 | p. Glu11* | stop codon | Hom | Proband | VUS |
| F9 | SGCB | c.416G>T | Exon 3 | p. Gly139Val | missense | Het | mother | VUS |
| F10 | SGCA | c.718T>A | Exon 3 | p. Gly240Val | missense | Hom | Proband | VUS |
| † The variants have been previously reported as pathogenic; ‡, novel variants; Hom, homozygous; Het, heterozygous; VUS Variant of Undetermined Significance, N/A not available. | ||||||||
Table 2: Genetic analysis of patients with sarcoglycanopathy.
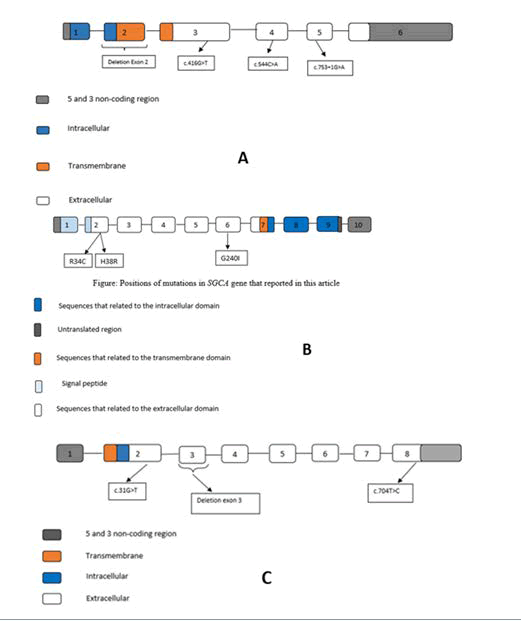
SGCA: We identified three missense mutations in SGCA, two of which have been reported previously [20, 21], and one was novel (c.100C>T (p. Arg34Cys), c.718T>A (p.Gly91Ser), c.113A>G (p.His38Arg)). Two mutations were located in exon 2, and one was in exon 1. All mutations were homozygote in patients with LGMD2D. (Table 2)
SGCB: Four mutations were identified in SGCA, which all of them have been previously reported [22]. Including two missense mutations c.544A>C (p.Thr182Pro) and c.416G>T (p.Gly139Val)), one deletion mutation (Exon 2 deletion), and one splicing mutation (c.753+1G>A). Except for the heterozygote mutation found in patient nine, other mutations were the homozygote state in LGMD2E patients.
SGCG: Three homozygote mutations were detected in SGCG; two of them have been already reported as a deletion and a missense mutation in ClinVar database (Exon 3 deletion and c.704T>C (p.Leu235Pro) respectively), and one was a novel stop codon mutation (c.31G>T (p.Glu11*)).
In 10 patients that we checked, there were no abnormalities on electrocardiographic or echocardiographic though generally, this group of genes is expressed in muscle and cardiac muscle.
Discussion
LGMD has been recognized as a broad and increasingly heterogeneous class of inherited muscle diseases [30]. With the increase in genetic discoveries of LGMD, it has become clear that clinical and histological manifestations and outcomes may vary widely between different subgroups and in affected individuals. However, these changes are not distinct and contradictory enough to enable physicians to identify new subtypes based on phenotype; hence, the diagnosis has always been challenging. With the appearance of NGS, the time and its cost required to sequence the entire human genome has reduced from years to days and from several billion dollars to several thousand dollars, respectively [31].
Today, due to the high number of genes involved in this disease, the approach of examining each gene by Sanger sequencing is not practical, and the next-generation sequencing, in which a set of genes can be examined simultaneously, is more cost-effective and time-consuming [32]. Currently, next-generation sequencing studies make it possible to analyze different types of genes simultaneously. However, these techniques must be performed carefully by adding clinical, radiological, and pathological data. The advent of next-generation sequencing approaches has accelerated the discovery of new LGMD genes. Ten years ago, the list included 16 loci, while today, the genes identified are thirty-one, with eight autosomal dominant and 23 autosomal recessive manners [32]. Conventional methods for identifying pathogenic mutations such as immunohistochemistry, Western blotting, and Sanger sequencing of selected genes can make a genetic diagnosis in 35% of families [33]. With the invention of Next Generation Sequencing (NGS), patients can now be screened through neuromuscular disease gene panels or by Whole Exome Sequencing (WES) [34,35].
Overall, it is reported that clinical exome sequencing has a 25% diagnosis rate [36]. In comparison, recent studies on exome sequencing for neuromuscular disease show a 46% diagnosis rate in the United States and 73% in the highly consanguineous population of Iran [37-38]. Although WES results require more validation for clinical diagnosis, in addition to the ability to screen all known disease-related genes, this method provides an additional opportunity to identify new genes associated with LGMD. Genetic diagnosis in LGMD and other genetic disorders will become increasingly important in the coming years as new molecular therapies target specific gene defects and even specific mutations. For example, several studies showed that LGMD2D is easily controllable by gene therapy approaches [39-41]. Any modifications that were found can disrupt the sarco glycan complex. All SG proteins are glycosylated membrane proteins with small intracellular and large extracellular domains. Four sarco glycan proteins form tetra meric membrane proteins fixed to the dystrophin axis by lateral association with the dystroglycan complex. Unlike dystroglycan, which is found in almost all cells, sarco glycans are predominantly found in muscle cells. Early studies in LGMD patients have shown that mutations in each of the four SG genes cause instability of the whole complex. However, further studies of patients and animal models have shown that not all sarco glycans are equally important for maintaining the stability of the complex.
In this study, we performed a comprehensive analysis of clinical phenotypes and genetic data in 10 sarco glycan-pathy-related patients from five separate provinces in Iran. Due to the locus heterogeneity in sarco glycanopathies, there is little information about its worldwide prevalence. However, reports that have the global prevalence of all forms of LGMD are estimated at one in 14,500 to 123,000[30]. Based on the given information, a different subtype of SGCs shows various frequencies. Generally, the prevalence of different types of sarcoglycanopathy varies according to ethnicity and geographical area. For example, LGMD2D is relatively common in Europe and the United States while LGMD2C is more common in India and Algeria populations. In a report by Nilipour et al. in 100 muscle samples biopsy from Iranian patients, it was shown that SGP is the second most common muscular dystrophy among other muscular dystrophies (for example, Duchenne muscular dystrophy) in Iran and the types of sarcoglycanopathy α-SGPs are the most abundant form [43-47].
Clinical features associated with SGC mutations in the Iranian group, including the age of onset, disease progression, presence/absence of specific features, and severity of symptoms, are very heterogeneous, even in patients with the same mutation phenotypic variation intrafamilial and interfamilial. This diversity has been reported previously and suggests that other genetic and environmental factors may be involved [48-49].
The rare cause of sarcoglycanopathy is Mutation in SGCB, and the frequency of patients with SGCB mutation varies from 5.5% in India to 23% in Brazil. In this study, four mutations in SGCB were reported, that one of these is a deletion of exon 2 in the homozygous state. According to ACMG guidelines, this mutation is pathogenic and, with the elimination of the anchor domain's SGCB protein, has destructive effects on the assembly of the sarcoglycan complex. The family that showed this mutation was from the southeast of Iran and the Balouch ethnic group. In another study by Alavi. It was found that approximately 85% (12 out of 14) of their LGMD2E patients have been detected, which showed the complete deletion of exon 2 in the SGCB gene, and 10 out of 12 families was from southeast of Iran too. Haplotype analysis supported by three Single Nucleotide Polymorphism Markers (SNPs) and high frequency of this mutation in this region indicated the probably founder effect in this region of Iran could be helpful in screening for mutations in patients diagnosed with LGMD2 in this area. We need to further study with more sample size and additional markers [48]. Different alleles are proved by haplotype analysis that is identical with the deletion mutation in SGCB by descent. Since almost all patients from southeastern Iran had this mutation, it is suggested that deletion of exon 2 should be the first target screening for patients with LGMD2D in this region. All of the modifications in the SGCB gene reported in this article are located in the extracellular domain of the SGCB gene, like the SGCA gene. All of the three said left mutations are in the extracellular environment.
Sequence change c.A544C in SGCB gene replaces threonine with proline at the protein level. Although threonine residue is a high conservative amino acid at this position, there is little difference between threonine and proline in terms of physicochemically. This variant was reported in the population database (rs751427686, ExAC 0.01%), and its frequency is in the range of mutations frequency. In another publication, various changes at this codon (p.Thr182Ala) have been found in the compound heterozygous state in affected individuals with a pathogenic variant on the SGCB gene, too (PMID: 9032047). According to databases that can predict the function and structure of the proteins, this change is probably damaging, damaging, disease-causing on databases SIFT, PROVEAN, MutationTaster, respectively. Still, these disruptive functions have not been approved by published functional studies. Therefore, this variant (c.A544C) was classified as Likely Pathogenic in ACMG databases and variant of unknown significance on Clinvar. In summary, we do not have enough information to find out the function of this variant.
Alpha is the most common type of sarcoglycanopathy [32-34]. The important function of sarcoglycan complex was first indicated with α-sarcoglycan (adrenaline) deficiency in patients with autosomal recessive muscular dystrophy in Arabic countries. Defects in the SGCs destroy the sarcolemma back-bone so that the cell membrane is exposed to contractile muscle pressures, and as a result, rupture of the focal membrane occurs. Eventually, the dystrophic phenotype appears [35-38]. In this study, three mutations in the SGCA gene were seen. SGCA gene mutations may prevent the formation of sarcoglycan complexes or the binding of sarcoglycan complexes to DGC. All three of these mutations are located in the extracellular domain of the protein. One of these mutations is c.100C>T. With this mutation, the codon encoding the arginine amino acid is converted to cysteine, located in the protein's extracellular domain, the N terminal of the protein. This domain binds alpha sarcoglycans to the extracellular matrix. These mutations are found in the cadherin-like domain of the SGCA. This domain is a location for many missense mutations that cause LGMD2D. The cadherin-like domain is located in the extracellular environment and is involved in protein-protein interlinkage, cell polarization, and migration. It also plays an important role in interdependence with other dystrophin-glycoprotein complex molecules. All missense mutations reported in this article are located in the extracellular domain of α-sarcoglycan and have a strong quantitative effect on protein expression levels, indicating that mutated regions are critical for the formation and stability of the sarcoglycan complex.
In addition to c.101G>A (p.Arg34His), another nucleotide substitution at the same codon c.100C>T (p.Arg34Cys) has been proven to cause LGMD. Remarkably, the missense mutations that happened in these sites, R34, D97, and R98, were mapped to the putative Ca2+ binding site in the cadherin-like domain [39-42]. These approved that these residues are crucial, and any mutations cause LGMD. However, there is no significant difference in clinical phenotype among patients with various mutations Arg34His, Arg34Cys, and Arg34Leu in the SGCA gene. The two rest mutations in SGCA (c.113A>G, c.718T>A) are novel, and we do not find any information on databases. Both were found in the homozygous state on probands and were heterozygous on all healthy members, such as healthy parents and siblings. In addition to that, both of these are damaging, damaging, and disease-causing on SIFT, PROVEAN, Mutation Taster databases. The gamma gene mutation was first described in the Maghreb African country of north Africa [43].
The variant c.704T>C is a new variant, and this missense mutation caused to replacement of amino acid Leucine with Proline. According to databases SIFT, PROVEAN, Mutation Taster, this variant is damaging and disease-causing [44].
Best regards to all patients participating in this study; Finally, it is suggested that a comprehensive cohort study of all multiple geographical regions and ethnicities should be carried out to obtain complete and accurate information on the abundance of genes and mutations in Iranian sarcoglycanopathy patients. Also, larger cohorts of patients need to define genotype/phenotype correlations in SGCs. Further studies can help us determine the frequency of different SGCs and mutations in the Iranian population. In rare diseases, the small population of patients prevents access to accurate and comprehensive clinical, genetic, and treatment information. To overcome this problem, it is recommended that reputable scientific centers worldwide work together and share their information, information that can be important and effective in clinical and genetic diagnosis and disease management and treatment[45-49].
Conclusion
In the present study, ten probands out of ten families suspected of being affected by LGMDs had a mutation (s) in SGCA, SGCG, and SGCB genes that four of them were novel. The most prevalent mutations of sarcoglycanopathy in our study were beta, and alpha and gamma were placed in the second stage. Missense's mutation was the most prevalent type, and in our research, no mutation was found in the SGCD gene.
Based on genetic classification four patients (%40) were affected with LGMD2E (SGCB mutations), three (%30) with LGMD2C (SGCG mutations), three (%30) with LGMD2D (SGCA mutations).
Our study could clarify the genetic cause of ten Iranian patients in ten unrelated families with ten different types of mutations. In this study, we found two new mutations that enrich human genetic mutation databases, and we were able to expand our knowledge about the genetic spectrum of LGMD in Iran. The present study expands the clinical and molecular spectrum of Sarcoglycanopathy related-genes in the study families. Our finding provided additional insights into genotype and phenotype correlations in the Iranian population.
References
- Nigro V, Aurino S, Piluso G (2011) Limb-girdle muscular dystrophies: update on genetic diagnosis and therapeutic approaches. Curr Opin Neurol 24:429-436
[Crossref] [Google scholar] [Indexed]
- Xie Z, Hou Y, Yu M, Liu Y, Fan Y, et al. (2019) Clinical and genetic spectrum of sarcoglycanopathies in a large cohort of Chinese patients. Orphanet J Rare Dis 14:43
[Crossref] [Google scholar] [Indexed]
- Araishi K, Sasaoka T, Imamura M, Noguchi S, Hama H, et al. (1999) Loss of the sarcoglycan complex and sarcospan leads to muscular dystrophy in β-sarcoglycan-deficient mice. Hum Mol Genet 8:1589-1598
[Crossref] [Google scholar] [Indexed]
- Alavi A, Esmaeili S, Nilipour Y, Nafissi S, Tonekaboni SH, et al. (2017) LGMD2E is the most common type of sarcoglycanopathies in the Iranian population. J Neurogenet 31:161-169
[Crossref] [Google scholar] [Indexed]
- Moore SA, Shilling CJ, Westra S, Wall C, Wicklund MP, et al. (2006) Limb-girdle muscular dystrophy in the United States. J Neuropathol Exp Neurol 65: 995-1003
[Crossref] [Google scholar] [Indexed]
- Tasca G, Monforte M, Díaz-Manera J, Brisca G, Semplicini C, et al. (2018) MRI in sarcoglycanopathies: a large international cohort study. J Neurol Neurosurg Psychiatry 89:72-77
[Crossref] [Google scholar] [Indexed]
- Dalichaouche I, Sifi Y, Roudaut C, Sifi K, Hamri A, et al. (2017) gamma-sarcoglycan and dystrophin mutation spectrum in an Algerian cohort. Muscle Nerve 56:129-135
[Crossref] [Google scholar] [Indexed]
- Khadilkar SV, Singh RK, Hegde M, Urtizberea A, Love DR, et al. (2009) Spectrum of mutations in sarcoglycan genes in the Mumbai region of western India: high prevalence of 525del T. Neurol India 57:406-410
[Crossref] [Google scholar] [Indexed]
- Lu Y, Song X, Ji G, Wu H, Li D, et al. (2019) Identification of a novel SGCA missense mutation in a case of limb-girdle muscular dystrophy 2D with the absence of four sarcoglycan proteins. Neuropathology 39:207-211
[Crossref] [Google scholar] [Indexed]
- Wicklund MP, Kissel JT (2014) The limb-girdle muscular dystrophies. Neurol Clin 32:729-749
[Crossref] [Google scholar] [Indexed]
- Politano L, Nigro V, Passamano L, Petretta V, Comi LI, et al. (2001) Evaluation of cardiac and respiratory involvement in sarcoglycanopathies. Neuromuscular Disorders 11:178-185
[Crossref] [Google scholar] [Indexed]
- Noguchi S, McNally EM, Ben Othmane K, Hagiwara Y, et al. (1995) Mutations in the dystrophin-associated protein gamma-sarcoglycan in chromosome 13 muscular dystrophy. Science 270:819-822
[Crossref] [Google scholar] [Indexed]
- Fannin M, Duggan DJ, Mostacciuolo ML, Freda MP Martinello F, et al., (1997) Genetic epidemiology of muscular dystrophies resulting from sarcoglycan gene mutations. Journal of medical genetics 34:973-977
[Crossref] [Google scholar] [Indexed]
- Bushby K, Beckmann J (2003) The 105th ENMC sponsored workshop: pathogenesis in the non-sarcoglycan limb-girdle muscular dystrophies, Naarden, April 12–14, 2002. Neuromuscul Disor13:80-90
[Crossref] [Google scholar] [Indexed]
- McDonald CM, Johnson ER, Abresch RT, Carter GT, Fowler WM Jr, et al. (1995) Profiles of neuromuscular diseases. Limb-girdle syndromes. Am J Phys Med Rehabil 74:S117-130
[Crossref] [Google scholar] [Indexed]
- Mojbafan M, et al. (2020) Mutational spectrum of autosomal recessive limb-girdle muscular dystrophies in a cohort of 112 Iranian patients and reporting of a possible founder effect. Orphanet J Rare Dis 15:14
[Crossref] [Google scholar] [Indexed]
- Ghafouri-Fard S, Bahmani R, Bagheri SD, Sharifi Z, Zeinali S (2017) Limb-Girdle Muscular Dystrophy Type 2E Due to a Novel Large Deletion in SGCB Gene. Iran J Child Neurol 11:57-60
- Mojbafan M, Hashemi-Gorji F, Fardaei M, Miryounesi M (2016) A novel mutation in alpha sarcoglycan gene in an Iranian family with limb-girdle muscular dystrophy 2D. Neurol Res 38:220-223
[Crossref] [Google scholar] [Indexed]
- Mojbafan M, Nilipour Y, Tonekaboni SH, Bagheri SD, Bagherian H, et al. (2016) A rare form of limb-girdle muscular dystrophy (type 2E) seen in an Iranian family detected by autozygosity mapping. J Neurogenet 30:1-4
[Crossref] [Google scholar] [Indexed]
- Carrie A, Piccolo F, Leturcq F, de Toma C, Azibi K, et al. (1997) Mutational diversity and hot spots in the alpha-sarcoglycan gene in autosomal recessive muscular dystrophy (LGMD2D). J Med Genet 34:470-475
[Crossref] [Google scholar] [Indexed]
- Park HJ, Jang H, Kim JH, Lee JH, Shin HY, et al. (2017) Discovery of pathogenic variants in a large Korean cohort of inherited muscular disorders. Clin Genet 91:403-410
[Crossref] [Google scholar] [Indexed]
- Duggan DJ, Gorospe JR, Fanin M, Hoffman EP, Angelini C (1997) Mutations in the sarcoglycan genes in patients with myopathy. N Engl J Med 336:618-624
[Crossref] [Google scholar] [Indexed]
- Guglieri M, Magri F, D'Angelo MG, Prelle A, Morandi L, et al. (2008) Clinical, molecular, and protein correlations in a large sample of genetically diagnosed Italian limb-girdle muscular dystrophy patients. Hum Mutat 29:258-266
[Crossref] [Google scholar] [Indexed]
- Arzani M, Rezaei H, Moghadasi AN (2018) Association of Limb-Girdle muscular dystrophy with multiple sclerosis: A case report. Caspian J Intern Med 9:96-99
[Crossref] [Google scholar] [Indexed]
- Nigro V, Savarese M (2014) Genetic basis of limb-girdle muscular dystrophies: the 2014 update. Acta myologica : myopathies and cardiomyopathies. Acta Myol 33:1-12
- Chu ML, Moran E (2018) The Limb-Girdle Muscular Dystrophies: Is Treatment on the Horizon? Neurotherapeutics. Neurotherapeutics 15:849-862
[Crossref] [Google scholar] [Indexed]
- Bansal D, Campbell KP (2004) Dysferlin and the plasma membrane repair in muscular dystrophy. Trends Cell Biol 14:206-213
[Crossref] [Google scholar] [Indexed]
- Beckmann JS, Spencer M (2008) Calpain 3, the "gatekeeper" of proper sarcomere assembly, turnover and maintenance. Neuromuscul Disord 18:913-921
[Crossref] [Google scholar] [Indexed]
- Astrea G, Pezzini I, Picillo E, Pasquariello R, Moro F, et al. (2016) TMEM5-associated dystroglycanopathy presenting with CMD and mild limb-girdle muscle involvement. Neuromuscul Disord 26:459-461
[Crossref] [Google scholar] [Indexed]
- Mitsuhashi S, Kang PB (2012) Update on the genetics of limb-girdle muscular dystrophy. Semin Pediatr Neurol 19:211-218
[Crossref] [Google scholar] [Indexed]
- Wang L, Ankala A, Al Khallaf H, Wang X, Martchenko M, et al. (2017) The Applications and Challenges of Next-Generation Sequencing in Diagnosing Neuromuscular Disorders 177-200
- Ku CS, Cooper DN, Polychronakos C, Naidoo N, Wu M, et al. (2012) Exome sequencing: dual role as a discovery and diagnostic tool. Ann Neurol 71:5-14
[Crossref] [Google scholar] [Indexed]
- Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, et al. (2009) Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461:272-276
[Crossref] [Google scholar] [Indexed]
- Yang Y, Muzny DM, Xia F, Niu Z, Person R, et al. (2014) Molecular findings among patients referred for clinical whole-exome sequencing. JAMA 312:1870-1879
[Crossref] [Google scholar] [Indexed]
- Ankara A, da Silva C, Gualandi F, Ferlini A, Bean LJ, et al. (2015) A Comprehensive Genomic Approach for Neuromuscular Diseases Gives a High Diagnostic Yield. Ann Neurol 77:206-214
[Crossref] [Google scholar] [Indexed]
- Kalhor Z, Fattahi Z, Fadaee M, Vazehan R, Parsimehr E, et al. (2016) Improved diagnostic yield of neuromuscular disorders applying clinical exome sequencing (ces) in patients arising from a consanguineous population. Clin Genet 91:386-402
- Pack CA, Walter GA, Gaidosh G, Bryant N, Lewis MA, et al. (2007) Long-term skeletal muscle protection after gene transfer in a mouse model of LGMD-2D. Mol Ther 15:1775-1781
[Crossref] [Google scholar] [Indexed]
- Mendell JR, Rodino-Klapac LR, Rosales XQ, Coley BD, Galloway G, et al. (2010) Sustained alpha-sarcoglycan gene expression after gene transfer in limb-girdle muscular dystrophy, type 2D. Ann Neurol 68:629-638
[Crossref] [Google scholar] [Indexed]
- Mendell JR, Rodino-Klapac LR, Rosales-Quintero X, Kota J, Coley BD, et al. (2009) Limb-girdle muscular dystrophy type 2D gene therapy restores alpha-sarcoglycan and associated proteins. Ann Neurol 66:290-297
[Crossref] [Google scholar] [Indexed]
- Trabelsi M, Kavian N, Daoud F, Commere V, Deburgrave N, et al. (2008) Revised spectrum of mutations in sarcoglycanopathies. Eur J Hum Genet 16:793-803
[Crossref] [Google scholar] [Indexed]
- Wicklund M, DiVincenzo C, Liaquat K, Karbassi I (2013) Relative Prevalence of Limb-Girdle Muscular Dystrophies in the United States Population (P07.030) AAN Enterprises.
- Meena A, Sreenivas D, Sundaram C, Rajasekhar R, Sita JS, et al. (2007) Sarcoglycanopathies: a clinico-pathological study. Neurol India 55:117-121
[Crossref] [Google scholar] [Indexed]
- McNally EM, Passos-Bueno MR, Bönnemann CG, Vainzof M, De Sá Moreira E, et al. (1996) Mild and severe muscular dystrophy caused by a single gamma-sarcoglycan mutation. Am J Hum Genet 59:1040-1047
- Sandonà D, Betto R (2009) Sarcoglycanopathies: molecular pathogenesis and therapeutic prospects. Expert Rev Mol Med 11:e28-e28
[Crossref] [Google scholar] [Indexed]
- Eymard B, Romero NB, Leturcq F, Piccolo F, Carrié A, et al. (1997) Primary adhalinopathy (alpha-sarcoglycanopathy): clinical, pathologic, and genetic correlation in 20 patients with autosomal recessive muscular dystrophy. Neurology 48:1227-1234
[Crossref] [Google scholar] [Indexed]
- Lo HP, Cooper ST, Evesson FJ, Seto JT, Chiotis M, et al. (2008) Limb-girdle muscular dystrophy: Diagnostic evaluation, frequency and clues to pathogenesis. Neuromuscul Disord 18:34-44
[Crossref] [Google scholar] [Indexed]
- Yamada H, et al. (2001) Processing of β-dystroglycan by matrix metalloproteinase disrupts the link between the extracellular matrix and cell membrane via the dystroglycan complex. Hum Mol Genet 10:1563-1569
[Crossref] [Google scholar] [Indexed]
- Ferreira AF, Carvalho MS, Resende MB, Wakamatsu A, Reed UC, et al. (2011) Phenotypic and immunohistochemical characterization of sarcoglycanopathies. Clinics 66:1713-1719
[Crossref] [Google scholar] [Indexed]
- NILIPOR Y, Shariatmadari F, Gorji, FA, Rouzrokh M, Ghofrani M, et al. (2013) Evaluation of one hundred pediatric muscle biopsies during a 2-year period in mofid children and toos hospitals. Iran J Child Neurol 7:17-21
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences