Biochemical Characterisation and Whole Genome Expression Profiling of Cultured Skin Fibroblasts from Two South African Adults with Urinary 3-Hydroxyisovaleric Acid and 3-Methylcrotonylglycine
Zandberg L, Erasmus E, Suormala T, Mienie LJ, Mels CMC and Van Dijk AA
DOI10.21767/2380-7245.100049
1Biochemistry, Focus Area Human Metabolomics, North-West University, Potchefstroom, South Africa
2Division of Metabolism, University Children's Hospital, Zurich, Switzerland
3Metabolic Unit, University Children's Hospital, Basel, Switzerland
4Hypertension in Africa Research Team, North-West University, Potchefstroom, South Africa
- *Corresponding Author:
- Van Dijk AA
Biochemistry Division, North-West University
Private Bag X6001 Noordbrug
2520 Potchefstroom, South Africa.
Tel: +27 18 299 2317
Fax: +27 18 299 2316
E-mail: albie.vandijk@nwu.ac.za
Received date: April 06, 2016; Accepted date: September 18, 2016; Published date: September 21, 2016
Citation: Zandberg L, Erasmus E, Suormala T, et al. Biochemical Characterisation and Whole Genome Expression Profiling of Cultured Skin Fibroblasts from Two South African Adults with Urinary 3-Hydroxyisovaleric Acid and 3-Methylcrotonylglycine. J Rare Disord Diagn Ther. 2016, 2:5. doi: 10.21767/2380-7245.100049
Copyright: © 2016 Zandberg L, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
We report on the first case of marginal 3-methylcrotonyl-CoA carboxylase (MCC) deficiency in South Africa. Urinary 3-hydroxyisovaleric acid and 3-methylcrotonylglycine were detected in four males of a non-consanguineous family. Only the index patient (NWU001) had non-specific symptoms, the others were asymptomatic. The inherited metabolite profile and partially reduced MCC activity were indicative of marginal MCC deficiency. In vivo L-leucine loading confirmed a reduced flux through the leucine degradation pathway. No known deleterious mutations were detected in the open reading frames of MCCC1 and MCCC2. NWU001 was heterozygous for a SNP in MCCC1 (rs2270968; c.1391A>C, p.H464P). NWU002 was heterozygous for a MCCC2 splice variant which skips exon 7 and causes an in frame deletion of 38 amino acids that is identical to a predicted shorter MCCC2 isoform-2 (Q9HCC0-2). Whole genome expression profiles from cultured skin fibroblasts of NWU001 and NWU002 and two healthy adults using Affymetrix® HuExST1.0 arrays detected 14237 significantly differentially expressed transcript IDs of which only 1277 have known annotation and gene association. The underlying molecular interactions, secondary signalling responses and functional relationships of these 1277 transcripts were inspected following a knowledge-based functional analyses approach using Ingenuity Pathway Analysis software. The transcriptome had a footprint of oxidative stress, disruption of energy homeostasis, inflammation, impaired cellular maintenance and repair mechanisms. Of note was the significant up regulation of the fatty acid amide hydrolase variant 2 (FAAH2) HuChrX transcript in the anandamide degradation canonical pathway. The observations that the two MCC transcripts were not significantly differentially expressed and that more than 90% of the significantly differently expressed transcripts are still poorly annotated further support the notion that secondary factors other than the MCC loci impact on the MCC deficiency phenome.
https://transplanthair.istanbul
https://hairclinicturkey.co
https://hairclinicistanbul.co
https://besthairtransplant.co
https://hairtransplantistanbul.co
Introduction
There is an increasing awareness that although inborn errors of metabolism are considered simple monogenic disorders caused by a deficiency of a single enzyme, they often act as complex diseases [1-5]. This is evident from the often broad spectrum of clinical symptoms and highly variable time of onset for the same disease [4,6-10]. One such disease is isolated 3-methylcrotonyl- CoA carboxylase (MCC) deficiency (MIM #210200 and MIM #210210) [11-14]. MCC deficiency is an autosomal recessive disorder of the leucine catabolism which usually presents with elevated levels of urinary 3-hydroxyisovaleric acid (HIVA) and 3-methylcrotonylglycine (MCG) [15-16]. Newborn screening (NBS) revealed that MCC deficiency is the most frequently detected organic aciduria [13,17,18]. MCC deficiency is regarded as a disease with a low penetrance [19] since less than 10% of individuals with a metabolite profile indicative of MCC deficiency develops [14]. The clinical, biochemical, and enzymatic characterization of MCC deficiency have been well documented, but the pathomechanism remains largely unknown [9,15,20-26].
MCC is one of four biotin-dependent carboxylases and is comprised of two non-identical subunits, alpha (MCCA) and beta (MCCB), which forms a α6β6 hetero-dodecamer [27-29]. The two subunits are encoded by nuclear genes and have signal peptides that target them to the mitochondrion [30]. The alpha subunit is encoded by the MCCC1 (formerly called MCCA) gene located on chromosome 3q25-q27 and has 19 coding exons. The beta subunit is encoded by the MCCC2 (formerly called MCCB) gene located on chromosome 5q12-q13 and has 17 coding exons [31- 33]. About 132 disease-causing mutations have been reported for the MCCC1 and MCCC2 genes [19] but no prevalent mutation has been identified and there is no clear genotype-phenotype correlation [17,21,31]. The MCC activity of patients with deleterious homozygous or compound heterozygous mutations is usually reduced to <5% of the mean activity of the controls [15,21,22,31], but a few cases have been reported with residual MCC activity, up to 34% [19,22,26] and some individuals with urinary metabolites indicative of MCC deficiency who only have a single heterozygous MCC mutation have almost normal MCC activity [17]. There is an increasing awareness of the difficulty to diagnose MCC deficiency because some patients present with an atypical metabolic profile with only trace amounts or even no detectable urinary MCG [22], the variability and non-specific nature of its symptoms and the growing number of individuals presenting with MCC deficiency associated metabolites but with only a single mutation in MCCC1 or MCCC2 [17]. Recently, analyses of the first exome sequence data from MCC deficiency cases [34] revealed that individuals who have the same mutation most likely share a common ancestor, that rare mutations outside of MCC genes can cause non-specific phenotypes hitherto attributed to MCC deficiency in genetically closely related patients and that the clinical consequences of MCC deficiency could be less pervasive than generally accepted. The authors also stated that further studies should be done to assess the value of treating asymptomatic individuals with MCC deficiency such as those who are identified through NBS programmes [34].
MCC deficiency has not yet been reported in South Africa. In this paper we present results from a non-consanguineous South African family of which four males presented with a metabolic profile with borderline high levels of HIVA and detectible amounts of MCG which suggest MCC deficiency. Only one of the affected males experienced non-specific symptoms since early adulthood, whereas the other three remained asymptomatic. Our mutation analyses of the open reading frames (ORF) of MCCC1 and MCCC2, enzymatic studies and in vivo L-leucine loading suggest that these individuals have marginal MCC deficiency. Intriguingly, the whole genome expression profile of cultured skin fibroblasts indicated that both MCCC1 and MCCC2 were not significantly differentially expressed in cells of the affected individuals compared to the controls. The gene expression profiling and inheritance pattern in this family seem to suggest that the chromosome-X (HuChrX) might impact on the pathomechanism of this disorder.
Clinical Presentation
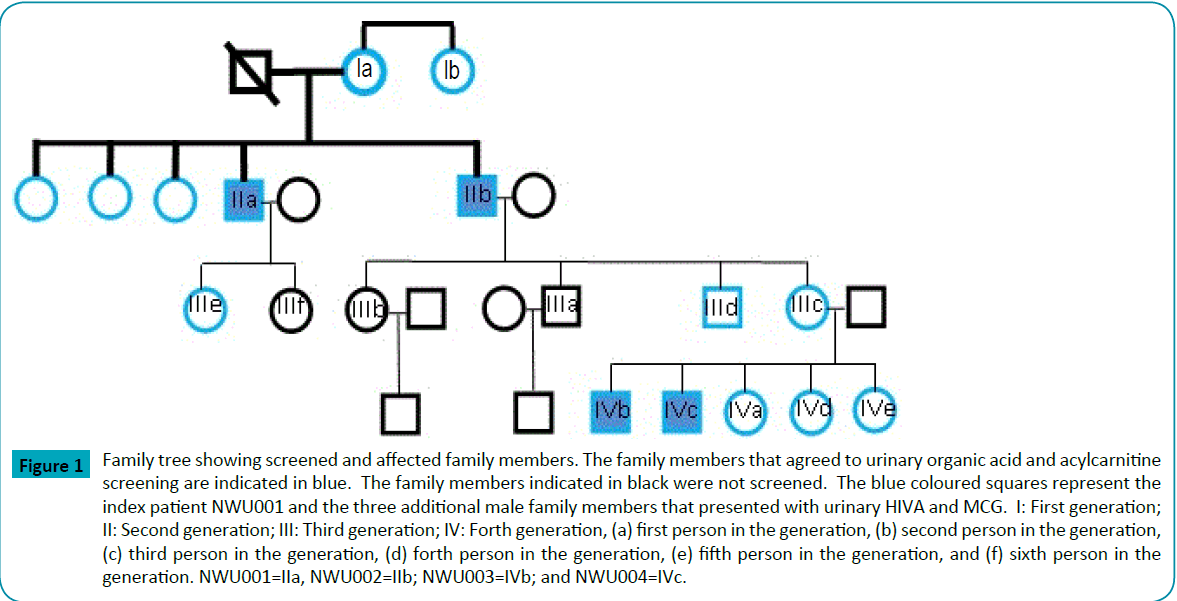
The index patient (NWU001), a Caucasian male from a nonconsanguineous South African family, was 48 years of age when he presented with mild hepatosplenomegaly. "The metabolic screen revealed elevated levels of urinary 3-hydroxyisovaleric acid and 3-methylcrotonylglycine which is usually indicative of MCC deficiency." He was advised to report for metabolic screening. The patient mentioned a 20 year history of frequently occurring symptoms often in association with episodes of chronic fatigue. He reported reduced tolerance to exercise and frequently experienced acute muscle cramps which subsided with rest. He complained of general weakness, poor tolerance to environmental factors and being prone to hay fever, eczema, mouth ulcers and sinusitis. Although the patient had a body mass index of 27.9 (Obese reference range: >26.9), he appeared healthy and not overweight. His blood chemistry was mostly within normal ranges, with the exception of a slightly elevated alanine aminotransferase activity of 41 U/L (Male age 12 months-60 years reference range: 10-40 U/L) and elevated levels of 784 pmol/L VitB12 (Normal reference range: 148 - 616 pmol/L). His total cholesterol was borderline high, 5.5 mmol/L (Borderline high reference range: 5.18-6.19 mmol/L) and he had low HDL, 0.4 mmol/L (Normal reference range: <1.04 mmol/L), normal LDL, 2.67 mmol/L (Normal reference range: <2.85 mmol/L) but very high levels of triglycerides, 6.73 mmol/L (normal reference range: <5.65 mmol/L) [35]. He presented with normal blood pressure, fasting glucose and haematological parameters.
The second family member, NWU002 (male, age 52), sibling of the index patient NWU001 had no clinical symptoms related to that of his brother. However, he also presented with a similar profile of abnormal urinary organic acids and acylcarnitine urinary metabolites as NWU001. At the time of investigation NWU002 already received medication to control persistent high levels of total cholesterol and his total cholesterol level was 4.9 mmol/L (Reference range: 2.8-5.2 mmol/L).
In addition to the index patient and his brother another thirteen family members over three generations (Figure 1), ten females (aged, 83, 80, 56, 53, 33, 7, and twins of 2 years) and three males (aged 31, 6 and 4 years), participated in a voluntarily metabolic screen. From the thirteen family members that were screened another two males in the family, the grandsons of NWU002 (NWU003, male, aged 6 years and NWU004, male, aged 4 years) also presented with a similar urinary organic acid profile as that of NWU001 and NWU002 (Supplementary Table 1). However, informed consent to include the grandsons (NWU003 and NWU004) for further testing could not be obtained and they were excluded from all further participation in this study.
Figure 1: Family tree showing screened and affected family members. The family members that agreed to urinary organic acid and acylcarnitine screening are indicated in blue. The family members indicated in black were not screened. The blue coloured squares represent the index patient NWU001 and the three additional male family members that presented with urinary HIVA and MCG. I: First generation; II: Second generation; III: Third generation; IV: Forth generation, (a) first person in the generation, (b) second person in the generation, (c) third person in the generation, (d) forth person in the generation, (e) fifth person in the generation, and (f) sixth person in the generation. NWU001=IIa, NWU002=IIb; NWU003=IVb; and NWU004=IVc.
Methods
Urinary metabolic screening
Creatinine concentration of the urine samples were determined colourimetrically with a modified Jaffe reaction. Urinary organic acids were extracted using liquid-liquid extraction and derivatization as decribed by Hoffman and Feyh [36]. Trimethylchlorosilane (TMCS) esters of organic acids were analysed on a Hewlett Packard 6890/5973 gas chromatography mass spectrometry (GC-MS) system equipped with a DB- 1ms capillary column (30 m × 0.25 mm × 0.25 μm, Agilent Technologies, Palo Alto) as described by Rinaldo [37]. The data was quantified using an automated mass spectral deconvolution and identification system (AMDIS) software. The concentration of identified organic acids were expressed in millimole per mole creatinine, using 4-phenylbutyric acid as internal standard.
Butylated urinary acylcarnitines were analysed according to the method described by Millington [38]. An Agilent 1200 series high pressure liquid chromatography system (Agilent Technologies, Palo Alto), equipped with a 96-well plate autosampler and a tripple quadrupole mass spectrometer was used (G6410, Agilent Technologies, Palo Alto). The system was operated in positive electrospray ionisation (ESI) mode with the mass analyzers set in a precursor of 85 m/z configuration. Acylcarnitines were quantified using deuterated analogues of free carnitine and acylcarnitines as internal standards.
In vivo leucine loading
NWU001, NWU002, and five control individuals (Aged between 24 and 64 years) participated in an L-leucine loading challenge. Each participant received 100 mg L-leucine (Sigma-Aldrich) per kg body weight in orange juice [39]. Urine samples were collected at baseline (T0) and 30 min (T1), 1 h (T2), 3 h (T3), 5 h (T4) and 7 h (T5) after L-leucine administration. Urinary organic acid and acylcarnitine analyses were conducted on all the samples.
Primary skin fibroblast cultures
Skin biopsies from NWU001 and NWU002 and three unrelated, healthy Caucasian adult control individuals (CON001, male aged 62 years, CON002, female aged 25 years and CON003, male aged 24 years) were obtained from the left inner forearm of each participant by a registered nurse. Primary skin fibroblast cultures were established by GenePath (Centurion, South Africa). Fibroblasts were routinely grown in T25 cm2 tissue culture flasks in Dulbecco's Modified Eagle’s Medium (DMEM, Lonza), supplemented with 10% foetal calf serum (FCS, Lonza), and 1% Penicillin/Streptomycin (Lonza) antibiotics at 37ºC under 5% CO2. NWU001, NWU002, CON001 and CON002 were also grown in Amniochrome system II® medium (Lonza) supplemented with 0,125 mg/ml Penicillin/Streptomycin (Lonza) and 0.256 mg/mL Gentamycin (Lonza) in T25 cm2 tissue culture flasks for total RNA isolation and hybridization to Affymetrix Genechip HuExST1.0 arrays. Cells were grown until they reached 80-90% confluence. The cell layers were washed twice with phosphate buffered saline (PBS) and total RNA was isolated directly from the flask.
Enzyme assays
Mitochondrial biotin dependent carboxylase activities were assayed in crude fibroblast lysates by measuring the incorporation of [14C]-NaHCO3 into acid-non-volatile products by established methods [40]. For the assay of the level of specific activity and kinetics of MCC, fibroblasts were grown in the routine culture medium supplemented with 10% FCS. This medium is biotin sufficient containing 9 nmol/L biotin, contributed by the natural biotin content of FCS, which is 5-times the mean normal plasma biotin concentration in humans (1.8 nmol/L, range 0.6-4.8, n = 126) [41,42]. The activity of another mitochondrial biotin dependent carboxylase, propionyl-CoA carboxylase (PCC) that serves as a control enzyme, was determined simultaneously with MCC in the same cell lysate. To determine the apparent Km values of MCC for the substrates 3-methylcrotonyl-CoA and bicarbonate as well as for ATP, MCC activity was measured with variable concentrations of one of the compounds together with routine saturating assay concentrations of the other components. Thus, the concentration of 3-methylcrotonyl-CoA varied between 0.05 and 3.9 mmol/L (in routine assay 1.5 mmol/L), Na-bicarbonate was between 0.42 and 40 mmol/L (in routine assay 20 mmol/L), and ATP was between 0.04 and 6.3 mmol/L (in routine assay 3.15 mmol/L). All Km values were derived using Eadie-Hofstee plots. To determine the sensitivity to the activator K+, MCC activity was measured with and without 50 mmol/L (optimal concentration) and 100 mmol/L KCl in the assay mixture.
Holocarboylase synthetase (HCS) activity was estimated indirectly by measuring the sensitivity of fibroblast MCC and PCC activities on biotin depletion of the culture medium [43]. Cells were grown in parallel in a medium with low and high biotin concentration and assayed for MCC and PCC activity. Low-biotin medium was prepared by replacing the FCS of the routine culture medium by newborn calf serum which resulted in the final biotin concentration of 0.1 nmol/L, and high-biotin medium by supplementing the low-biotin medium with 10 μmol/L biotin. Biotinidase activity was estimated in serum by the colourimetric assay using N-biotinyl-p-aminobenzoate as substrate [44].
Mutation analysis of MCCC1 and MCCC2
Total RNA was extracted using the Qiazol® reagent (Qiagen) and the RNeasy® total RNA extraction Kit (Qiagen). cDNA was synthesised from total RNA isolated from cultured skin fibroblasts of NWU001 and NWU002 using cloned AMV reverse transcriptase (Invitrogen). Phusion® high fidelity DNA polymerase (Finnzymes) was used to amplify MCCC1 and MCCC2 from cDNA using published primer sequences [31,33]. All amplification products were purified with the QiaQuick® gel extraction kit (Qiagen) and sequenced (Inqaba Biotech). The MCCC1 and MCCC2 cDNA sequences were inspected for mutations using FinchTV 1.4.0 (Geospiza, Inc.; Seattle, WA, USA; https://www.geospiza.com) and aligned to reference sequence NM_020166 for MCCC1 and NM_022132 for MCCC2 using the web based Clustal W (Version 2.1) multiple sequence alignment tool [45].
Whole genome gene expression profiling
HuExST1.0 array preparation and hybridisation: The whole genome expression profile was determined with the Affymetrix® GeneChip® Human Exon ST 1.0 (HuExST1.0) array for hybridisation (Affymetrix). Total RNA was isolated from fibroblasts of NWU001 and NWU002 and two healthy control adults (CON001 and CON002) using the RNeasy® mini kit (Qiagen) with no additional DNase digestion. The yield, quality and integrity of the total RNA preparation was assessed according to the integrity of the 28S rRNA and 18S rRNA, as well as RNA integrity number (RIN) value using a NanoDrop 1000 Spectrophotometer and an Agilent Bioanalyser 2100. Single hybridisations of NWU001, NWU002 and the controls, CON001 and CON002, were prepared. Hybridisation was carried out by the Affymetrix® service provider in South Africa, the Centre of Proteomic and Genomic Research (CPGR), Cape Town. Total RNA from each sample with a RIN >9 was fragmented, labelled, hybridised to HuExST1.0 arrays and scanned according to the Affymetrix® protocols and instructions. Output signal intensity data files (.CEL) were generated and assessed to ensure good quality output and analyses.
HuExST1.0 array data analysis and significantly differentially expressed transcript ID-list: The data intensity files (.CEL files) received from CPGR were analysed using Affymetrix® Expression Console (Version 1.1, HuEx1-0-st-v2.na32.hg19) and Partek® Genomic Suite software (Version 6.1, Copyright, Partek Inc. St. Louis, MO, USA). Pre-processing data quality control analyses were done by using the robust multi-array average (RMA) normalisation method for background correction and using the core-meta probe set as annotation for quality control analyses [46,47]. All data intensity files (.CEL) passed the QC metric and were uploaded with Partek® GS software (Version6.1, Copyright, Partek Inc. St. Louis, MO, USA). The Affymetrix® full-meta probe set (HuEx1-0-st-v2.na32.hg19) was applied for annotation. The full-meta probe set included transcript IDs with well-defined annotations as well as predicted transcripts with no known gene associations. A list of differentially expressed, annotated and predicted transcript IDs was generated by means of one way analysis of variance (ANOVA). The significantly differentially expressed transcript list included the transcript IDs that met the filter criteria P<0.05. From the significantly differentially expressed list, a subset of well-annotated transcript IDs associated with transcripts associated with genes of the HuChrX was also compiled.
Functional analyses
Ingenuity Pathway Analysis (IPA) software (Ingenuity® Systems, www.ingenuity.com, build version 242990, content version 17199142), was applied to investigate the predicted functional relationships, predicted network associations, biological functions, canonical pathways, regulator-, co-activator- and modifier genes associated with the complete list of annotated differentially expressed transcript IDs. A subset of significantly differentially expressed well-annotated transcript IDs associated with HuChrX was compiled and also analysed with IPA (Ingenuity® Systems, www.ingenuity.com, build version 313398M, content version 18841524) to further investigate the possible impact of the HuChrX transcripts on the functional relationships, network associations, biological functions, canonical pathways and regulatory networks of this marginal MCC deficiency. The analysis settings for IPA for both input lists were set to include only well annotated transcript IDs and endogenous chemicals identified in the predefined IPA database with a predicted direct and or indirect relationship in Homo sapiens.
Quantitative real-time PCR validation
Independent quantitative real-time PCR (qPCR) analysis was performed to confirm the gene expression results of selected transcripts obtained from the HuExST1.0 array hybridisations. The same total RNA samples were used for both the array hybridisation and qPCR analyses. An amount of 1 μg total RNA of each sample was reverse transcribed using Cloned-AMV reverse transcriptase (Invitrogen). The cDNA samples were diluted and 100 ng was used in each of the validation reactions which were prepared in triplicate. TaqMan® hydrolysis probe chemistry based gene expression assays for ACOX2 (Hs00185873_m1), AWAT2 (Hs01040631_m1), HADHA (Hs00426191_m1), HMGCL (Hs00609311_m1), NDUFS2 (Hs00190020_m1) and UXT (Hs00188238_m1) were selected. Amplification was done as proposed by the manufacturer using the ABI 7500 system. Quantification cycle (Cq) values were determined for each sample using the comparative Cq method. Relative gene expression ratios were normalized to the reference genes 18S rRNA and hypoxanthine phosphoribosyltransferase 1 (HPRT1). Relative mRNA expression was represented as the fold change of the target group (marginal MCC deficiency) over the calibrator sample (control). The calibrator sample was defined as the mean expression of samples and replicates of the control group. Relative gene expression changes were calculated according to the 2-ΔΔCt method [48,49].
Results
Urinary metabolic profiling
The metabolic screen of index patient NWU001 revealed an abnormal urinary organic acid profile with a borderline high normal level of HIVA and detectable amounts of MCG. No secondary metabolites associated with the precursor molecule, propionyl-CoA was present. The urinary acylcarnitine profile complemented the organic acid profile and revealed slightly elevated levels of 3-hydroxyisovalerylcarnitine (C5OH) and 3-methylcrotonylcarnitine (C5.1), but no severe secondary carnitine deficiency [36,50]. This patient seemed to be the first South African (Caucasian) reported with possible MCC deficiency. Molecular characterisation and further investigation was approved by the ethics committee of the North-West University (Approval number 05M 14). Fifteen family members over three generations (Figure 1), participated in a metabolic screen. Similar abnormal urinary organic acids and acylcarnitine urinary profiles as NWU001 were detected in three additional males in the family, the brother (NWU002) of NWU001 and grandsons of NWU002 (NWU003 and NWU004) (Supplementary Table 1).
In vivo L-leucine loading test
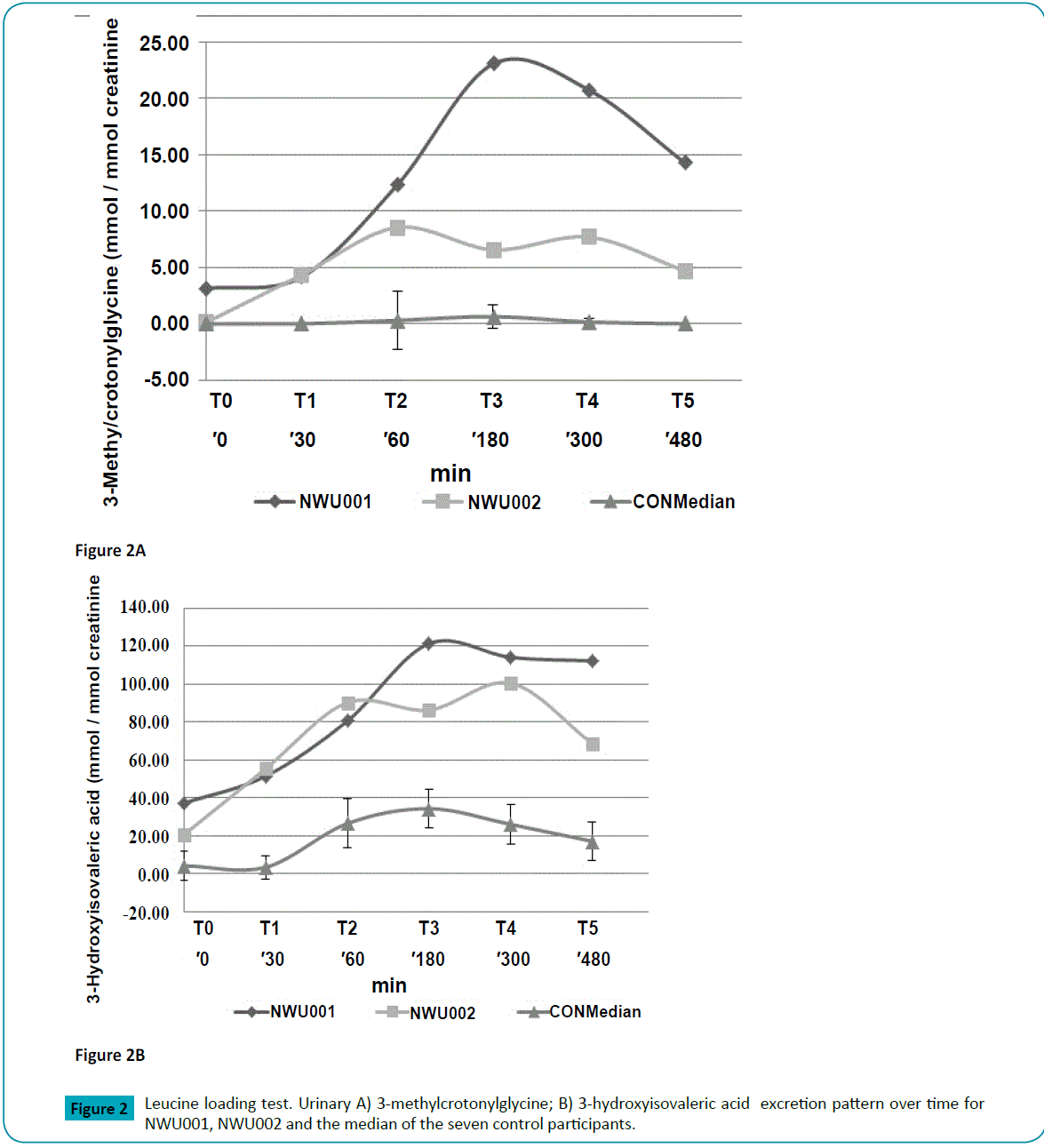
To further investigate the leucine catabolism, an in vivo L-leucine loading test was performed comparing the metabolite profile of the leucine degradation pathway of NWU001 and NWU002 and five healthy Caucasian adult controls. The loading test confirmed that NWU001 and NWU002 had elevated baseline levels of the urinary HIVA and MCG (Figure 2; Supplementary Table 2). After L-leucine administration, a distinct elevation of HIVA and MCG was observed in NWU001 and NWU002. The maximum level of HIVA for NWU001 was measured at T3, approximately three times higher at T3 than at T0 (T0, 37.1 and T3, 121.3 mmol/mol creatinine), whereas NWU002 had a maximum elevation of HIVA at T4, almost five times higher at T4 than at baseline (T0, 20.5 and T4, 100.4 mmol/mol creatinine). For NWU001, the levels of MCG increased more than seven times, reaching a maximum at T3 (T0, 3.1 and T3, 23.1 mmol/mol creatinine). For NWU002 the level of MCG increased almost nine times reaching a maximum at T2 (T0, 0.10 and T2, 8.6 mmol/mol creatinine). Between T4 and T5 after commencement of the L-leucine challenge, the excretion levels of both HIVA and MCG of the two patients, NWU001 and NWU002, started to decrease. The administration of L-leucine did not affect the metabolic flux of the control participants. For the controls, the maximum level of excreted urinary HIVA was 32 mmol/mol creatinine measured at T2, which was still much lower than the baseline of NWU001. A very slight change in MCG levels in the control individuals occurred which quickly resolved, whereas NWU001 and NWU002 had detectable levels of MCG that persisted for the duration of the seven hours of monitoring.
Enzyme activities
Specific activities of MCC and PCC in fibroblasts grown in the standard FCS-based medium obtained in three or four replicate experiments are shown in Table 1. In fibroblasts of NWU001 and NWU002 MCC activities were slightly below or just within the reference range with mean activities reduced to 40% and 36% of the mean reference value, respectively, whereas PCC activities were within the reference range. Marginally reduced MCC activity in cells of NWU001 and NWU002 was confirmed when MCC activity was correlated with PCC activity assayed in the same cell lysate. Thus, in these two cell lines PCC/MCC activity ratios were elevated or just at the upper reference range. In contrast, MCC and PCC activities as well as the PCC/MCC activity ratios were well within the reference range in the three simultaneously assayed control cell lines.
| Cell line* | Carboxylase activities, pmol/min/mg protein | Ratio | ||||
|---|---|---|---|---|---|---|
| MCC | PCC | PCC/MCC | ||||
| Individual values** | Mean | Individual values** | Mean | Individual values | Mean | |
| NWU001 | 80.2, 96.8, 134, 216 | 132 | 209, 323, 483, 558 | 393 | 2.60, 3.34, 3.60, 2.58 | 3.03 |
| NWU002 | 76.0, 95.1, 135, 165 | 118 | 286, 452, 535, 586 | 465 | 3.76, 4.75, 3.97, 3.76 | 4.01 |
| CON001 | 329, 345, 492 | 389 | 641, 673, 1042 | 786 | 1.95, 1.95, 2.12 | 2.01 |
| CON002 | 205, 243, 244 | 231 | 356, 493, 516 | 455 | 1.73, 2.03, 2.12 | 1.96 |
| CON003 | 352, 404, 566 | 440 | 490, 776, 831 | 699 | 1.39, 1.92, 1.47 | 1.59 |
| Reference values, range and mean | ||||||
| 53 different cell lines | 134–671 | 328 | 208–1165 | 617 | 1.19–2.58 | 1.95 |
Table 1: Activities of 3-methylcrotonyl-CoA carboxylase (MCC) and propionyl-CoA carboxylase (PCC) in crude lysates of fibroblasts grown in the standard FCS-supplemented biotin sufficient medium.
Kinetic studies (Table 2) revealed that the Km values of MCC for 3-methylcrotonyl-CoA, Na-bicarbonate and ATP as well as the requirement for the activator K+ in cells of NWU001 and NWU002 were not importantly different from those obtained in the three simultaneously assayed control cell lines. These results exclude reduced affinity of MCC for the substrates, ATP and activator K+, as the cause for the marginally reduced activity of MCC in cells of the patients.
| Cell lines* | Km for 3-MC-CoA μmol/L | Km for Na-bicarbonate mmol/L | Km for ATP μmol/L | MCC activity, % of optimal** | |
|---|---|---|---|---|---|
| No K+ | 100 mmol/L K+ | ||||
| NWU001 | 332 | 4.26 | 87.5 | 23 | 85.5 |
| NWU002 | 560 | 3.49 | 117 | 25 | 87.5 |
| CON001 | 513 | 4.11 | 128 | 20.6 | 83.7 |
| CON002 | 705 | 3.55 | 116 | 21.1 | 88.9 |
| CON003 | 388 | 4.03 | 94.4 | 16.9 | 93 |
Table 2: Km values of 3-methylcrotonyl-CoA carboxylase (MCC) for its substrates 3-methylctotonyl-CoA (3-MC-CoA) and Na-bicarbonate as well as for ATP, and the effect of varying the concentration of the activator K+ measured in crude fibroblast lysates.
Results in Table 3 show that the sensitivity of the fibroblasts of NWU001 and NWU002 to biotin depletion of the culture medium was not increased. Thus, after one passage in low-biotin medium MCC activity, which is more sensitive to biotin depletion than PCC activity, was reduced to 28-44% of that in high-biotin medium in cells of the patients which is similar to that seen in the three simultaneously assayed control cell lines and within the reference range. These findings exclude mild HCS deficiency as the cause for the abnormalities in the patients. However, the mean MCC activity in fibroblasts of NWU001 and NWU002 grown in the high-biotin medium was marginally reduced to 34% and 29% of the mean reference value, respectively, i.e., to similar levels as obtained in cells grown in the FCS-based medium.
| Carboxylase activities | pmol/min/mg protein | ||||
|---|---|---|---|---|---|
| Biotin in medium** | MCC activity*** | %**** | PCC activity*** | %**** | |
| NWU001 | Low | 36.2, 55.4 | 28, 45 | 181, 398 | 49, 68 |
| High | 129, 124 | 371, 586 | |||
| NWU002 | Low | 30.5, 46.5 | 37, 34 | 129, 202 | 50, 45 |
| High | 82.2, 136 | 256, 452 | |||
| CON001 | Low | 76.5, 119 | 15, 23 | 405, 547 | 35, 52 |
| High | 525, 520 | 1172, 1045 | |||
| CON002 | Low | 54.3 | 22 | 189 | 39 |
| High | 249 | 485 | |||
| CON003 | Low | 140, 160 | 36, 29 | 377, 646 | 57, 56 |
| High | 385, 557 | 663, 1155 | |||
| Reference values (30 different cell lines) | |||||
| Mean | Low | 203 | 54 | 550 | 77 |
| Range | Low | 64.5-577 | 17-94 | 217-1094 | 33-102 |
| Mean | High | 376 | - | 727 | - |
| Range | High | 163-708 | - | 348-1514 | - |
**Fibroblasts were grown simultaneously for one passage in a medium supplemented with 10% new-born calf serum and a final biotin concentration of 0.1 nmol/L (low) and in the same medium supplemented with 10 μmol/L biotin (high);
***Values are the mean activities of duplicate determinations from replicate experiments;
****Activity in low-biotin medium , % of activity in simultaneously assayed activity in high-biotin medium
Table 3: Activities of 3-methylcrotonyl-CoA carboxylase (MCC) and propionyl-CoA carboxylase (PCC) in crude lysates of fibroblasts grown in media with low and high biotin concentrations.
The possibility of biotinidase deficiency was also excluded since the biotinidase activity of NWU001 in serum was 6.2 nmol/min/ mL which is within the reference range of 3.5 -12 nmol/min/mL [43].
Mutation analyses of the open reading frames of MCCC1 and MCCC2
Sequencing of MCCC1 and MCCC2 cDNA of NWU001 and NWU002, detected no known deleterious homozygous or compound heterozygous mutations. However, in MCCC1 of NWU001 a heterozygous SNP (rs2270968; c.1391A>C, p.H464P) was identified and one of the MCCC2 alleles of NWU002 contained a splice variant which resulted in the skipping of exon 7 to cause an in frame deletion of 38 amino acids (p.del209-I-A-VV- M-G-S-C-T-A-G-G-A-Y-V-P-A-M-A-D-E-N-I-I-V-R-K-Q-G-T-I-F-LA- G-P-P-L-246) resulting in a 525 amino acid peptide. This second peptide sequence had 100% sequence similarity to a predicted MCCC2 isoform-2 (XM_005248624.1, Q9HCC0-2). No second mutation to suggest a homozygous or compound heterozygous genotype was detected in the ORF of either MCCC1 or MCCC2 of NWU002.
Whole genome expression profiling
The significantly differentially expressed transcript ID list of the marginally affected MCC deficient fibroblasts consisted of 14237 transcript IDs (Supplementary Table 3), of which more than 90% (13328 transcript IDs) currently have no known or only poorly defined gene associations and functional annotation. Only 1277 transcript IDs, less than 10%, have known annotation and gene association (Supplementary Table 4) and were included in further functional analyses. Importantly, the two MCC transcripts, MCCC1 and MCCC2, were not significantly differentially expressed in the marginal MCC deficient transcriptome. To study the functional relationships, predicted network associations, biological functions, canonical pathways and toxicological associations between the 1277 well annotated transcript IDs in the dataset, IPA software (build version 242990, content version 17199142) was used.
Since the inheritance pattern of the metabolite profile in this South African family suggested a possible association with the HuChrX, the 14237 significantly differentially expressed transcript IDs were also sorted according to their chromosome association. Approximately 10% of transcript IDs have known and 90% have unknown annotations and gene associations for any particular chromosome. Of the 1277 well annotated significantly differentially expressed transcript IDs, 48 were associated with genes of HuChrX. Functional analysis of this subset was also performed separately (Supplementary Table 5).
Functional networks, the genetic footprint and possible implications of the marginally affected MCC deficient skin fibroblast transcriptome
The underlying molecular interactions, secondary signalling responses and functional relationships observed in the marginally affected MCC deficient skin fibroblast transcriptome were inspected following a knowledge-based functional analyses approach using IPA software. The 1277 well-annotated significantly differentially expressed transcripts indicated changes in functional networks associated with nucleic acid metabolism, cell-to-cell signalling and interaction, molecular transport, as well as cellular function and maintenance. A significant number of transcripts seemed to be involved with functional networks associated with the development of cancer (57%, 730/1277, P = 3.83 × 10-18) and gastrointestinal disease (21%, 268 /1277, P = 2.18 × 10-4). Only a relatively small number of canonical pathways were significantly affected. The canonical pathway, trigger receptor expressed on myeloid cells (TREM1) was one of the canonical pathways that had a considerable number of differentially expressed transcripts in this transcriptome. TREM1 belongs to the immunoglobin (Ig) family of cell surface receptors [51]. Activation of TREM1 triggers pathways leads to phosphorylation of signal transducers of transcription and nuclear factor kappa beta (NF-kB), which could trigger pro-inflammatory immune responses [52]. Transcripts of the granzyme A signalling and D-myo-inositol (1,3,4)-triphosphate biosynthesis pathways were also differentially expressed. D-myo-inositol (1,3,4)-triphosphate is an important secondary messenger molecule known to mediate signal transduction [53] of a large number of hormones, neurotransmitters, lipid signalling as well as regulating calcium release from intracellular stores [53]. Furthermore, more than 17 upstream regulators and a significant number of their target transcripts were significantly differentially expressed. The most significantly represented upstream regulators and their associated target transcripts in the dataset were E2F (11 target molecules, P = 3.40 × 10-4), ATF3 (4 target molecules, P = 4.94 × 10-3), MAPK1 (15 target molecules, P = 2.12 × 10-2), NRXN1 (5 target molecules, P = 2.15 × 10-2) RNU7-1 (2 target molecules, P = 2.47 × 10-2) and ELAVI (7 target molecules, P = 2.64 × 10-2). The interactome of the top affected predicted upstream regulators, their association with each other and their significantly differentially expressed target transcripts were inspected (Supplementary Figure 1).
Functional analyses and targeted inspection of pathways and networks of interest
The marginal MCC deficiency transcriptome was further characterised by a targeted approach of inspecting selected pathways of interest overlaid with the significantly differentially expressed transcripts. The significantly differentially expressed transcripts associated with branched chain amino acid degradation, glycolysis, gluconeogenesis, tricarboxylic acid (TCA) cycle, oxidative phosphorylation system (OXPHOS), fatty acid oxidation, ketogenesis, eicosanoid signalling and xenobiotic metabolism are listed in Table 4. This targeted approach resulted in some key observations for the characterisation of the pathomechanism associated with marginal MCC deficiency. Only one L-leucine degradation pathway associated transcript (HMGCL) was significantly down regulated. The HMGCL gene encodes 3-hydroxy-3-methylglutaryl-CoA lyase, which catalyses the last step of L-leucine catabolism. This enzyme (HMGCL) also catalyses the first step in the ketogenesis pathway, the process by which ketone bodies are produced as a result of fatty acid oxidation. Two other transcripts HADHA and ALDH6A1 associated with proteins of branched chain amino acid degradation pathways were also significantly down regulated. The secondary transcriptional response also seemed to impact the glycolysis/gluconeogenesis pathway as a slight induction of GPI and ME3 and down regulation of MDH1B was observed. One TCA associated transcript, ACO1, was up regulated, whereas three other transcripts, MDH1B, DHTKD1, and SDHD of the same pathway were down regulated. Three complexes of the OXPHOS system were affected, the NDUFS2 of complex I and SDHD of complex II were down regulated, while two complex IV associated transcripts MT-CO1/COX1 and COX6B were up regulated, and COX17 was down regulated. The fatty acid oxidation-I pathway was also affected with the down regulation of BDH2 and, as already mentioned, HADHA and HMGCL. Seven transcripts of the eicosanoid signalling pathway were differentially expressed. Three transcripts, PLA2G2A, PLA2G7 and PTGER4 were down regulated and three transcripts PLA2G4E, GGT1 and PTGIR, were induced. Several transcripts of the xenobiotic metabolism signalling pathway specifically associated with the aryl hydrocarbon receptor (AHR) ligand activation, PXR ligand/CAR agonist activation, and MEKK signalling cascade were significantly differentially expressed (Table 4). Affected transcripts of the AHR activation pathway include RELA, HSPA5, PTGES3, ANKRA2, and ALDH1A2. Affected PXR ligand/CAR agonist activation transcripts include CYP2C18, CYP2B6, SULT1E1, HS3ST1 and MRP3/ABCC3. Affected MEKK signalling cascade transcripts include MAP3K and GCLC.
| CanonicalPathway | Gene name/description | GeneSymbol | PValue | Foldchange | * | EntrezID |
|---|---|---|---|---|---|---|
| Branched chain amino aciddegradation | ||||||
| Isoleucinedegradation | Hydroxyacyl-CoAdehydrogenase/3-ketoacyl-CoAthiolase/enoyl-CoA hydratase (trifunctional protein), alphasubunit |
HADHA | 4.17E-02 | -1.39 | ## | 3030 |
| Leucinedegardation | 3-hydroxymethyl-3-methylglutaryl-CoAlyase | HMGCL | 1.99E-02 | -1.13 | ## | 3155 |
| Valinedegradation | Hydroxyacyl-CoAdehydrogenase/3-ketoacyl-CoAthiolase/enoyl-CoA hydratase (trifunctional protein), alphasubunit |
HADHA | 4.17E-02 | -1.39 | ## | 3030 |
| Aldehyde dehydrogenase 6 family, memberA1 | ALDH6A1 | 4.64E-02 | -1.29 | ## | 4329 | |
| Cholestrerolbiosynthesis | ||||||
| Sterol-C5-desaturase | SC5DL | 4.19E-02 | -1.26 | ## | 6309 | |
| Eicosanoidbiosynthesis | ||||||
| Phospholipase A2, groupIVE | PLA2G4E | 2.76E-03 | 1.10 | 123745 | ||
| Phospholipase A2, group IIA (platelets, synovialfluid) | PLA2G2A | 6.89E-03 | -1.31 | ## | 5320 | |
| Phospholipase A2, group VII (platelet-activating factoracetylhydrolase, plasma) |
PLA2G7 | 1.18E-02 | -1.09 | ## | 7941 | |
| Gamma-glutamyltransferase1 | GGT1 | 1.72E-02 | 1.32 | 2678 | ||
| Prostaglandin I2 (prostacyclin) receptor(IP) | PTGIR | 2.84E-02 | 1.23 | 5739 | ||
| Prostaglandin E receptor 4 (subtypeEP4) | PTGER4 | 4.44E-02 | -1.35 | ## | 5734 | |
| Fatty acid alphaoxidation | ||||||
| Aldehyde dehydrogenase 1 family, memberA2 | ALDH1A2 | 1.22E-02 | 1.21 | 8854 | ||
| Fatty acid betaoxidation | ||||||
| Hydroxyacyl-CoAdehydrogenase/3-ketoacyl-CoAThiolase/enoyl-CoA hydratase (trifunctional protein), alphasubunit |
HADHA | 4.17E-02 | -1.39 | ## | 3030 | |
| Gluconeogenesis | ||||||
| Malic enzyme 3, NADP(+)-dependent,mitochondrial | ME3 | 8.19E-03 | 1.07 | 10873 | ||
| Glucose-6-phosphateisomerase | GPI | 3.02E-02 | 1.18 | 2821 | ||
| Malate dehydrogenase 1B, NAD(soluble) | MDH1B | 4.77E-02 | -1.11 | ## | 130752 | |
| Glycolises | ||||||
| Glucose-6-phosphateisomerase | GPI | 3.02E-02 | 1.18 | 2821 | ||
| Heat Shock Proteininteraction | ||||||
| DnaJ (Hsp40) homolog, subfamily C, member5 | DNAJC5 | 7.00E-03 | 1.20 | 80331 | ||
| Heat shock 70kDa protein 5 (glucose-regulated protein,78kDa) | HSPA5 | 4.91E-02 | 1.69 | 3309 | ||
| Heat shock 60kDa protein 1(chaperonin) | HSPD1 | 6.15E-03 | -1.64 | ## | 3329 | |
| Kinesinfamily member20B | KIF20B | 2.11E-02 | -1.20 | ## | 9585 | |
| N-ethylmaleimide-sensitivefactor | NSF | 1.97E-02 | -1.11 | ## | 4905 | |
| Proteasome (prosome, macropain) 26S subunit, ATPase,2 | PSMC2 | 4.14E-03 | -1.44 | ## | 5701 | |
| Prostaglandin E synthase 3(cytosolic) | PTGES3 | 2.81E-02 | -1.75 | ## | 10728 | |
| Toll-like receptor3 | TLR3 | 4.68E-02 | -1.36 | ## | 7098 | |
| Toll-like receptor5 | TLR5 | 2.97E-02 | 1.99 | 7100 | ||
| Toll-like receptor6 | TLR6 | 3.95E-02 | -1.62 | ## | 10333 | |
| Ketogenesis | ||||||
| 3-hydroxymethyl-3-methylglutaryl-CoAlyase | HMGCL | 1.99E-02 | -1.13 | ## | 3155 | |
| 3-hydroxybutyrate dehydrogenase,type2 | BDH2 | 3.47E-02 | -1.59 | ## | 56898 | |
| Hydroxyacyl-CoAdehydrogenase/3-ketoacyl-CoAthiolase/enoyl-CoA hydratase (trifunctional protein), alphasubunit |
HADHA | 4.17E-02 | -1.39 | ## | 3030 | |
| Oxidativephosphorylation | ||||||
| Cytochrome c oxidase subunitI | COX1 | 2.04E-02 | 1.34 | 4512 | ||
| COX17 cytochrome c oxidase copperchaperone | COX17 | 3.07E-02 | -2.17 | ## | 10063 | |
| Succinate dehydrogenase complex, subunit D, integral membraneprotein | SDHD | 3.29E-02 | -1.17 | ## | 6392 | |
| Cytochrome c oxidase subunit VIb polypeptide 1(ubiquitous) | COX6B1 | 3.97E-02 | 1.31 | 1340 | ||
| NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49kDa(NADH-coenzyme Qreductase) |
NDUFS2 | 4.83E-02 | -1.22 | ## | 4720 | |
| Tricyclic acidcycle | ||||||
| Dehydrogenase E1 and transketolase domain containing1 | DHTKD1 | 3.12E-02 | -1.16 | ## | 55526 | |
| Succinate dehydrogenase complex, subunit D, integral membraneprotein | SDHD | 3.29E-02 | -1.17 | ## | 6392 | |
| Aconitase1,soluble | ACO1 | 4.48E-02 | 3.59 | 48 | ||
| Malate dehydrogenase 1B, NAD(soluble) | MDH1B | 4.77E-02 | -1.11 | ## | 130752 | |
| Xenobioticmetabolismsignaling | ||||||
| AHRsignaling | Aldehyde dehydrogenase 1 family, memberA2 | ALDH1A2 | 1.22E-02 | 1.21 | 8854 | |
| Aldehyde dehydrogenase 6 family, memberA1 | ALDH6A1 | 4.64E-02 | -1.29 | ## | 4329 | |
| Ankyrinrepeat, family A (RFXANK-like),2 | ANKRA2 | 1.95E-02 | -1.15 | ## | 57763 | |
| Heat shock 70kDa protein 5 (glucose-regulated protein,78kDa) | HSPA5 | 4.91E-02 | 1.69 | 3309 | ||
| Prostaglandin E synthase 3(cytosolic) | PTGES3 | 2.81E-02 | -1.75 | ## | 10728 | |
| v-rel avian reticuloendotheliosis viral oncogene homologA | RELA | 4.83E-03 | -1.32 | ## | 5970 | |
| TEBP | ||||||
| CAR-PXRsignaling | ATP-binding cassette, sub-family C (CFTR/MRP), member3 | ABCC3/MRP3 | 4.08E-02 | 1.17 | 8714 | |
| Cytochrome P450, family 2, subfamily B, polypeptide6 | CYP2B6 | 4.23E-02 | 1.21 | 1555 | ||
| Cytochrome P450, family 2, subfamily C, polypeptide18 | CYP2C18 | 1.26E-02 | -1.19 | ## | 1562 | |
| Sulfotransferasefamily 1E, estrogen-preferring, member1 | SULT1E1 | 4.24E-02 | -1.84 | ## | 6783 | |
| Heparansulfate (glucosamine) 3-O-sulfotransferase1 | HS3ST1 | 4.01E-02 | -1.36 | ## | 9957 | |
| MEKKsignaling | Glutamate-cysteine ligase catalyticsubunit | GCLC | 2.31E-02 | -1.31 | ## | 2729 |
| Mitogen-activated protein kinase kinasekinase6 | MAP3K6 | 9.60E-03 | -1.21 | ## | 9064 | |
Table 4: Important canonical pathways and associated significant differentially expressed transcripts in the marginal 3-methylcrotonyl-CoA carboxylase deficient skin fibroblast transcriptome.
Heat shock proteins (HSP) interact with the xenobiotic metabolism signalling via interaction through the AHR ligand activation pathway. Since HSP function was previously suggested as a possible secondary factor that could cause reduced enzyme activity of carboxylases [54], the expression of HSP was also inspected. The significantly differentially expressed transcripts of the heat shock protein interaction network were DNAJC5, HSPA5, HSPD1, KIF20B, NSF, PSMC2, PTGES3, TLR3, TLR5, and TLR6 (Supplementary Figure 2).
Since oxidative stress and unregulated reactive oxygen species (ROS) were reported to contribute to MCC deficiency related neurological symptoms [55-57], the knowledge-based ROS interactome of marginal MCC deficiency was compiled and inspected. Secondary signalling responses and the associated impact on target transcripts of the interactome were identified with IPA software (Figure 3). The ROS interactome suggested activation of the NADPH oxidase complex which is associated with increased ROS production. The activated NADPH oxidase complex and elevated ROS levels also impact on the pro-inflammatory cytokine regulatory system which involves the regulation of PLA2, NFκβ and Hsp70 interaction networks. The persistent activation of NADPH oxidase and increased ROS production seemed to cause chronic activation of TNF-α and other immune and inflammatory related response pathways.
Functional analysis of the HuChrX transcript subset
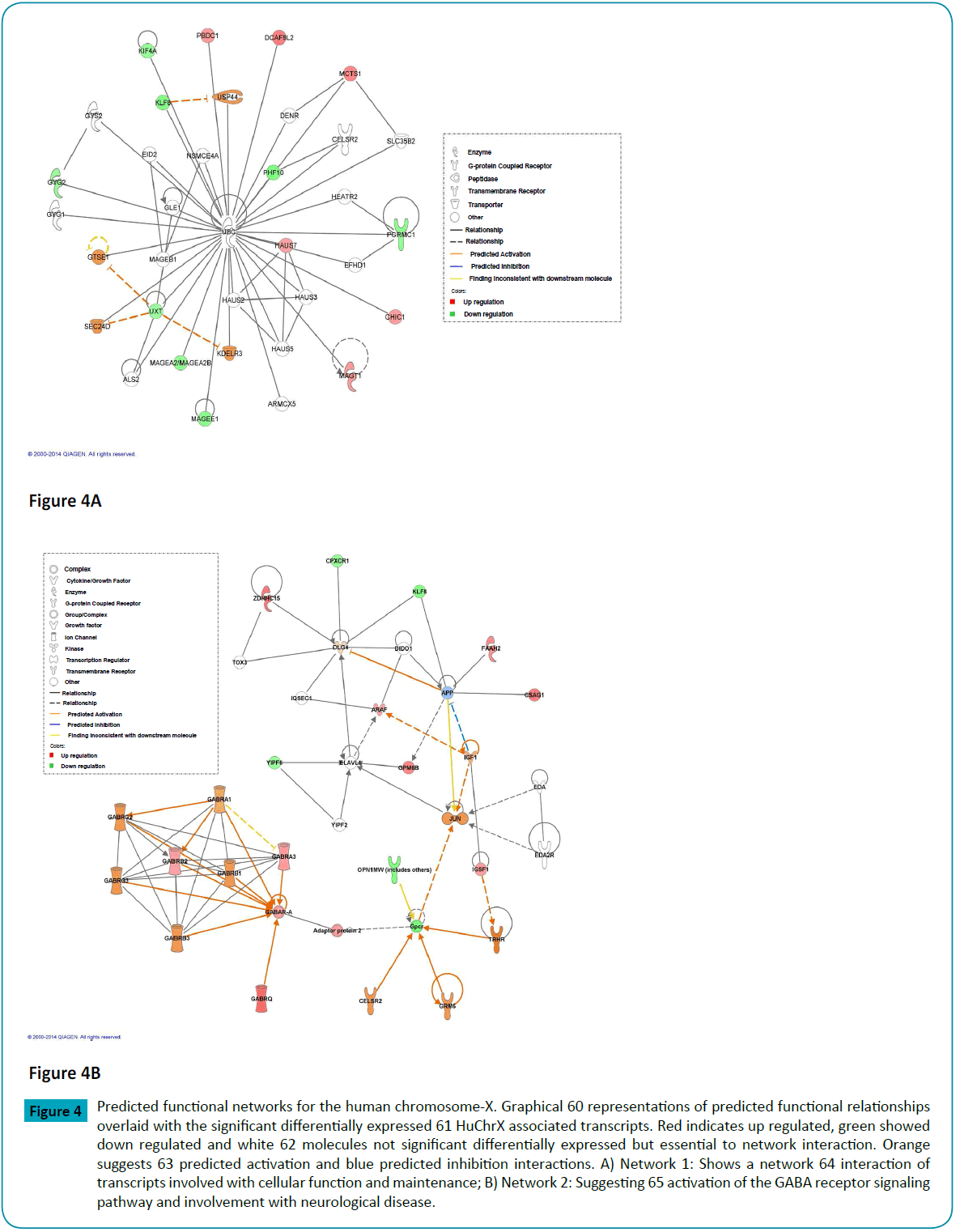
To better understand the specific contribution of HuChrX associated transcripts to the marginal MCC deficiency transcriptome, the subset of 48 significantly differentially expressed HuChrX associated transcripts were inspected separately. Functional network analysis showed that these 48 HuChrX associated transcripts clustered mainly in two independent functional networks (Figures 4a and 4b; Supplementary Table 6). The first network included molecules functionally important for molecular and cellular functions that involve cellular assembly, organisation, optimal function, and maintenance (P = 5.03 × 10-4 - 4.74 × 10-2). The second functional network predominantly included molecules involved with neurological development (P = 6.52 × 10-4 - 4.74 × 10-2) and lipid metabolism (P = 1.51 × 10-3 - 9.05 × 10-3). The most affected canonical pathways indicated by the HuChrX transcripts were GABA receptor signalling, anandamide degradation and glycogen biosynthesis II from UDP-D glucose pathways. One of the most interesting findings of the differentially expressed HuChrX associated transcript analyses was the 1.28 fold induction of the FAAH2 transcript. FAAH2 is one of two transcripts that code for fatty acid amide hydrolases, the other transcript is FAAH. These enzymes convert fatty acid amides to ethylamine and the corresponding fatty acid. While FAAH seems to prefer poly-unsaturated fatty acid amides as substrates, it seems as if FAAH2 is more specific for mono-unsaturated fatty acid amides [58]. However, FAAH2 is not yet well characterised. It is important to note that induction of FAAH/FAAH2 has been reported to be associated with pro-inflammatory signalling [59].
Independent qPCR validations
The HuExST1.0 array gene expression data was validated with independent qPCR analysis of eight selected significantly differentially expressed transcripts, HADHA, HMGCL, ACOX2, AWAT2, NDUFS2 and UXT using TaqMan hydrolysis probe chemistry gene expression assays. The fold change of the six selected differentially expressed transcripts from the HuExST1.0 array experiment was compared to the fold change obtained with the independent qPCR analyses. The data from triplicate experiments were analysed and the expression ratios of genes between the two groups (marginal MCC deficiency and Controls) were calculated using the 2-ΔΔCt method [48,49]. The expression profiles of these two independent datasets confirmed one another (Supplementary Figure 3).
Discussion
Here we present results of an investigation of the first evidence of MCC deficiency detected in four males in South Africa. The metabolite profile, indication of heritability, enzyme studies and in vivo L-leucine loading suggested that they have marginal MCC deficiency. Overall, the biochemical, enzymatic and genetic profile of NWU001 and NWU002 seemed similar to that of a group of previously described patients with partially reduced MCC activity [17]. The increased PCC/MCC ratio for NWU001 and NWU002 was similar to that reported for patients with elevated metabolites indicative of MCC deficiency and only one mutation in either MCCC1 or MCCC2 [17,19]. Sequencing of the coding region revealed that NWU001 and NWU002 did not share a common MCC genotype. NWU001 and NWU002 only presented with one heterozygous SNP and did not have a second mutation in either MCCC1 or MCCC2. These results are similar to a previous report wherein it was suggested that only one mutant allele in the presence of a wild type allele could also possibly explain the biochemical phenotype indicative of MCC deficiency [17]. Follow up studies are needed to further investigate the functional characteristics of the shorter MCCC2 isoform-2 detected in NWU002, which lacks exon 7, to better understand the impact of this heterozygous genetic variation.
In addition to the usual biochemical work-flow, we generated a transcriptome from cultured skin fibroblasts of the two marginal MCC deficient South African patients, NWU001 and NWU002, and two control adult individuals. We inspected the differentially expressed transcripts and secondary signalling responses for clues to the pathomechanism that might affect the MCC deficiency phenome. Two important observations from this transcriptome support the notion that secondary factors other than the MCC loci impact on the presentation of MCC deficiency. Firstly, both MCC transcripts, MCCC1 and MCCC2, were not significantly differentially expressed. This is in contrast to the transcriptome of symptomatic, clinical severely affected cases of MCC deficiency where MCCC1 and MCCC2 were significantly differentially expressed. Secondly, the finding that the vast majority (>90%) of the significantly differently expressed transcripts are still poorly annotated also seem to suggest a role of as yet unknown transcripts in this disorder.
Although the number of arrays analysed in this study was very limited and could raise concern for over interpretation and bias, the underlying molecular interaction networks and associated differentially expressed transcripts confirm and extend results from various previous studies on MCC deficiency. Similar to the transcriptome of symptomatic, clinically severe MCC deficiency and neurological studies on the cerebral cortex of young rats [55-57,60-65], this marginal MCC deficiency skin fibroblast transcriptome also had a genetic footprint suggestive of partial mitochondrial dysfunction, oxidative stress and disruption of energy homeostasis. Transcripts of mitochondrial respiratory chain complexes I, II and IV were significantly differentially expressed (Table 4) affecting the OXPHOS system (NDUFS2, SDHD, MT-CO1/COX1, COX6 and COX17). This result correlates with the finding that the metabolites, MCG and MCA, inhibited mitochondrial respiratory chain complexes II and III in vitro in rat brain homogenates [55] and could induced lipid and protein oxidative damage [55,57,64].
Aconitase 1 (ACO1), a hypoxia and oxidative stress sensitive transcript associated with TCA, was induced, an indication of elevated ROS and increased oxidative stress [66,67]. The NADPH oxidase complex was also induced, another indication of increased hydrogen peroxide, superoxide, hydroxyl radical, and oxygen radicals production and elevated levels of peroxynitrite [68]. Figure 3 shows the relationship between the induced NADPH oxidase activity, increased ROS production and activation of the pro-inflammatory responses via the eicosanoid biosynthesis pathway [68,69]. The induction of the NADPH oxidase complex usually induces NFκβ signalling [68], but this seems not to be the case in this transcriptome since RELA, a transcript coding for a protein of the NFκβ complex was down regulated and thus seem to regulate the NFκβ signalling pathway. This suppression of the NFκβ complex also impacts on the aryl hydrocarbon receptor (AHR) signalling cascade of the xenobiotic metabolism signalling and regulates the expression of genes involved in the innate and adaptive immune response.
Figure 3: Graphic representation of the marginal MCC deficiency ROS interactome. Graphical representation of the implicated ROS interactome overlaid with the significantly differentially expressed transcripts. Red indicates up regulated, green down regulated and white molecules not significant differentially expressed but essential to network interaction. Orange suggests predicted activation and blue predicted inhibition interactions.
Several transcripts that affect the AHR activation pathway and HSP networks were significantly differentially expressed. The significantly altered HSP network included the Hsp70 network (HSPA5) and ATPase (HSPD1), but not transcripts of Hsp60, which were reported to possibly contribute to phenotypic differentiation in MCC deficiency [54,70]. The HSP interaction network (Supplementary Figure 2) also showed significant differentially expressed Toll-like receptors which could further influence the inflammatory signalling as well as activation of the NFκβ and TNF α inflammatory signalling cascades that can add to the oxidative stress burden [71,72]. This marginal MCC deficiency ROS interactome suggests chronic activation of inflammatory and immune signalling responses. Chronic activated of pro-inflammatory signalling has been associated with compromised cellular membrane integrity and the development of cancer [69,73], Parkinson’s disease [74-76], Alzheimer’s disease [74,75,77-80], rheumatoid arthritis [59,81] and cardiovascular disease [61,82]
Both the complete set of 1277 and the subset of 48 HuChrX associated differentially expressed transcript IDs indicated increased inflammatory signalling responses and impaired cellular maintenance and repair activity that could imply compromised membrane integrity (Figure 4a; Supplementary Table 6). It is important to note that persistent oxidative stress in addition to an already compromised membrane repair capacity could severely affect membrane integrity. Membrane integrity homeostasis is highly regulated and a complex machinery that impact on lipid and bioactive lipid signalling through the activation of a phospholipase interaction network [59]. A striking and a perhaps important observation that could aid elucidating the underlying X-linked contribution to marginal MCC deficiency, was the significant up regulation of the FAAH2 (fatty acid amide hydrolase variant 2), a transcript of the anandamide degradation canonical pathway. The control of FAAH/FAAH2 expression was reported to have promising therapeutic value to control inflammatory related clinical symptoms [59]. Induction of the FAAH2 transcript implies a possible activation of the hydrolase activity converting anandamide to arachidonic acid (AA) and ethanolamide and thus contributes to the activation of the biosynthesis of proinflammatory bioactive lipids [59]. The overexpression of fatty acid amide hydrolases can compromise innate immunity in Arabidopsis [83]. Furthermore, inhibition of FAAH/FAAH2 seems to normalize cardiovascular function in hypertensive rats as well as normalise endotoxin-induced enhanced gastrointestinal motility in mice [84]. Strong associations between aberrant regulation of FAAH/FAAH2 and alcohol and substance dependency, as well as obesity [85] have been made. The overexpression of FAAH/FAAH2 also associates with sleep deprivation [58] as well as cholesterol level maintenance [86]. Incidentally, the index patient NWU001 experienced a number of symptoms which seem to correlate with the transcriptome footprint and abovementioned literature. Another reason for the specific interest in FAAH2 with regard to the unravelling of the underlying molecular pathomechanism of MCC deficiency is that NWU001 and NWU002 both had notably high blood triglycerides levels but only slightly reduced HDL, normal LDL and borderline elevated cholesterol levels. This might be a coincidence, but if not, could be an indicator to predict and monitor the development of disease and should be further investigated.
Figure 4: Predicted functional networks for the human chromosome-X. Graphical 60 representations of predicted functional relationships overlaid with the significant differentially expressed 61 HuChrX associated transcripts. Red indicates up regulated, green showed down regulated and white 62 molecules not significant differentially expressed but essential to network interaction. Orange suggests 63 predicted activation and blue predicted inhibition interactions. A) Network 1: Shows a network 64 interaction of transcripts involved with cellular function and maintenance; B) Network 2: Suggesting 65 activation of the GABA receptor signaling pathway and involvement with neurological disease.
A number of phospholipase family 2 (PLA2) transcripts were also differentially expressed. These enzymes have multiple functions in membrane phospholipid maintenance, homeostasis and the production of lipid mediators [87]. The PLA2 family releases fatty acids from the sn-2 position of the membrane phospholipids [88]. Since the sn-2 position of phospholipids consists mainly of AA, docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA), fatty acids are released to the cell [80] by the activation of specific PLA2 enzymes. Increased cellular AA enables the bioactive lipid signalling metabolism to synthesise pro-inflammatory eicosanoids [87] or be recycled into phospholipid by an acyltransferase [89]. The PLA2 family consists of six subtypes of which cytosolic PLA2 (cPLA2) is AA specific, whereas calcium-independent PLA2 (iPLA2) seems to be DHA specific [80,82]. The marginal MCC deficiency transcriptome, however, showed differential expression of the cPLA2 transcripts PLA2G2A, PLA2G4E, and PLA2G7 which suggests inhibition of the cPLA2 machinery, probably in an attempt to regulate the pro-inflammatory signalling. A prolonged inflammatory state causes cell damage and is associated with the pathogenesis of many diseases [90,91]. Also, cPLA2 seems to play a major role in lipid class switching [92] and is therefore considered as an important decision maker for regulating inflammation. The acute inflammatory process stays unresolved and develops into chronic inflammation when the pro-resolving mediators are not formed or are not functioning correctly. Significant differential expression of other eicosanoid biosynthesis pathway related transcripts GGT, PTGIR, PTGER4 and PTGES3 was also observed. The character of these transcripts predicted activation of PTGES2 (cyclooxygenase-2, COX-2), which suggests activation of the lipid mediator biosynthesis pathway. The enzymes encoded by PTGES genes are both pro- and anti-inflammatory lipid mediators. However, the induction of GGT suggests elevated levels of leukotriene D4 and the suppression of PTGES3 suggests elevated levels of prostacyclin which both have strong pro-inflammatory properties [93]. Imbalances in eicosanoid metabolism signalling have been detected in individuals with a weak tolerance to environmental factors and susceptibility to environmental stressors that often result in asthma [94-97], allergy, eczema [98-100], arthritis [81] as well as the development of cancer [73] and might also play a role in the development of non-specific symptoms of marginal MCC deficiency. Therefore, life-long monitoring of ROS levels of individuals with MCC deficiency along with the administration of anti-oxidants to prevent and manage deleterious secondary signalling responses may be beneficial as previously proposed (Figure 3) [55,56,60,64,65,101].
The apparent X-linked inheritance of urinary HIVA and MCG in this family was intriguing because MCC deficiency is an autosomal recessive inherited disease and there are no reports on an X-linked association. Another indication of a possible HuChrX-link in MCC deficiency was the report wherein six of eight patients who presented with metabolites of MCC deficiency, but for whom no MCC mutations could be detected were males [17]. The incidence of 3-methylglutaconic aciduria Type-I, another autosomal recessive disorder of the leucine catabolism, also seems to be higher in males than in females [16,102]. The apparent HuChrX link in both MCC and MCGAType- I deficiencies could be coincidental, but should be further investigated. Independent analyses of the subset of 48 HuChrX transcripts showed a significant impact on the GABA interactome. Although GABA receptors and signalling are tissue-specific, the activation and suppression of some of the GABA receptors specific for muscle function, organ and brain health in this skin fibroblast transcriptome and could possibly underlie some of the non-specific, mild psychological and muscle related symptoms experienced by the index patient NWU001. Aberration of GABA signalling pathways is well characterised for neuron cells and associated with the development of psychological disease [103].
Upstream regulators with the most significant overlap between their target molecules and the significantly differentially expressed transcripts were closely inspected. Interestingly, several downstream target genes of these regulatory factors are located on HuChrX. The upstream regulator E2F had the most significant representation of target transcripts in this transcriptome, two of which are located on HuChrX. The progesterone receptor membrane component 1 (PCRMC1) is a protein binding transmembrane receptor that regulates cholesterol amongst other molecules. Another HuChrX encoded target molecule of the upstream E2F transcription regulator is the ubiquitously-expressed, prefolding-like chaperone (UXT). This protein functions as a cofactor that modulates androgen receptor dependent transcription (Supplementary Figure 1).
Conclusion
In conclusion, this first investigation of marginal MCC deficiency in South Africa was initiated when NWU001 and three other male family members presented with urinary HIVA and MCG. The marginal MCC deficiency skin fibroblast transcriptome had a footprint of oxidative stress, partial mitochondrial dysfunction, disruption of energy homeostasis, inflammation, and impaired cellular maintenance and repair mechanisms, similar to what was observed in the transcriptome of clinically severe MCC deficient skin fibroblasts [104]. The functional analyses indicated that although MCC deficiency is considered a non-disease with little clinical impact, the secondary signalling responses predict a chronic activation of pro-inflammatory signalling and ROS production which could result in a detrimental long-term effect and should therefore be controlled. The observations that the two MCC transcripts, MCCC1 and MCCC2, were not significantly differentially expressed and that more than 90% of the significantly differently expressed transcripts are still poorly annotated, makes it impossible to come to firm conclusions and further support the notion that secondary factors other than the MCC loci impact on the MCC deficiency phenome.
Acknowledgements
We thank Prof MR Baumgartner for valuable discussions and critical reading of the manuscript. This study was funded by the Centre for Human Metabonomics, North-West University, Potchefstroom, the South African Department of Science and Technology (BioPAD, BPP007) and the South African National Research Foundation (Grant FA2005031700015).
Conflict of Interest
Lizelle Zandberg, Lardus Erasmus, Carina Mels, Terttu Suormala, Japie Mienie, and Alberdina van Dijk declare that they have no conflict of interest.
Informed Consent
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000 (5). Informed consent was obtained from all patients for being included in the study.
References
- DippleKM, McCabe ERB(2000) Modifier Genes Convert “Simple” Mendelian Disorders to Complex Traits. Mol Gen Metab 71: 43-50.
- Lachmann RH, Murphy EM (2014)The Adult Patient with Hereditary Metabolic Disease.In: Beaudet AL, Vogelstein B, Kinzler KW, Antonarakis SE, Ballabio A, et al. (eds.) The Online Metabolic and Molecular Bases of Inherited Disease. The McGraw-Hill Companies, Inc., New York, USA.
- Lanpher B,Brunetti-Pierri N, Lee B (2006) Inborn errors of metabolism: the flux from Mendelian to complex diseases. Nat Rev Genet 7: 449-460.
- Scriver CR (2007) The PAH gene, phenylketonuria, and a paradigm shift. Hum Mutat 28: 831-845.
- Touw CM, Derks TG, Bakker BM, Groen AK, Smit GP, et al. (2014) From genome to phenome-Simple inborn errors of metabolism as complex traits. BiochimBiophysActa 1842: 2021-2029.
- Gibson KM, Bennett MJ, Naylor EW, Morton DH(1998) 3-Methylcrotonyl-coenzyme A carboxylase deficiency in Amish/Mennonite adults identified by detection of increased acylcarnitines in blood spots of their children. J Pediatr 132: 519-523.
- Lee PJ (2002) Growing older: the adult metabolic clinic. J Inherit Metab Dis 25: 252-260.
- Sedel F, Baumann N, Turpin JC, Lyon-Caen O, Saudubray JM, et al. (2007) Psychiatric manifestations revealing inborn errors of metabolism in adolescents and adults. J Inherit Metab Dis 30: 631-641.
- Visser G, Suormala T, Smit GP, Reijngoud DJ, Boelkens B, et al.(2000) 3-methylcrotonyl-CoA carboxylase deficiency in an infant with cardiomyopathy, in her brother with developmental delay and in their asymptomatic father. Eur J Pediatr 159: 901-904.
- Walter J (2007) IEMs in adults. J Inherit Metab Dis 30: 627.
- Arnold GL, Koeberl DD, Matern D, Barshop B, Braverman N,et al. (2008) A Delphi-based consensus clinical practice protocol for the diagnosis and management of 3-methylcrotonyl CoA carboxylase deficiency. Mol Genet Metab 93: 363-370.
- Wilcken B, Wiley V, Hammond J, Carpenter K (2003) Screening newborns for inborn errors of metabolism by tandem mass spectrometry. N Engl J Med 348: 2304-2312.
- Lam C, Carter JM, Cederbaum SD, Neidich J, Gallant NM, et al. (2013) Analysis of cases of 3-methylcrotonyl CoA carboxylase deficiency (3-MCCD) in the California newborn screening program reported in the state database. Mol Genet Metab 110: 477-483.
- Stadler SC, Polanetz R, Maier EM,Heidenreich SC, Niederer B, et al.(2006)Newborn screening for 3-methylcrotonyl-CoA carboxylase deficiency: population heterogeneity of MCCA and MCCB mutations and impact on risk assessment. Hum Mutat 27: 748-759.
- Sweetman L, Williams JC(2001) Branched Chain Organic Acidurias.In: Scriver CRBA, Sly WS Valle D (eds.) The metabolic and molecular basis of inherited disease. McGraw-Hill, New York, pp: 2125-2163.
- Vockley J, Zschocke J, Knerr I,Vockley CW, Michael Gibson K(2014) Branched Chain Organic Acidurias.In: Beaudet AL, Vogelstein B, Kinzler KW, Antonarakis SE, Ballabio A, et al. (eds.) The Online Metabolic and Molecular Bases of Inherited Disease. The McGraw-Hill Companies, Inc., New York, USA.
- Morscher RJ, Grünert SC, Bürer C, Burda P, Suormala T, et al. (2012) A single mutation in MCCC1 or MCCC2 as a potential cause of positive screening for 3-methylcrotonyl-CoA carboxylase deficiency. Mol Genet Metab 105: 602-606.
- Feuchtbaum L, Carter J, Dowray S, Currier RJ, Lorey F (2012) Birth prevalence of disorders detectable through newborn screening by race/ethnicity. Genet Med 14: 937-945.
- Grunert SC, Stucki M, Morscher RJ, Suormala T, Burer C, et al. (2012) 3-methylcrotonyl-CoA carboxylase deficiency: clinical, biochemical, enzymatic and molecular studies in 88 individuals. Orphanet J Rare Dis 7: 31.
- Baumgartner MR (2005) Molecular mechanism of dominant expression in 3-methylcrotonyl-CoA carboxylase deficiency. J Inherit Metab Dis 28: 301-309.
- Dantas MF, Suormala T, Randolph A, Coelho D, Fowler B, et al. (2005) 3-Methylcrotonyl-CoA carboxylase deficiency: mutation analysis in 28 probands, 9 symptomatic and 19 detected by newborn screening. Hum Mutat 26: 164.
- Wolfe LA, Finegold DN, Vockley J, Walters N, Chambaz C, et al. (2007) Potential misdiagnosis of 3-methylcrotonyl-coenzyme A carboxylase deficiency associated with absent or trace urinary 3-methylcrotonylglycine. Pediatrics 120: e1335-e1340.
- Baykal T, Gokcay GH, Ince Z, Dantas MF, Fowler B, et al. (2005) Consanguineous 3-methylcrotonyl-CoA carboxylase deficiency: early-onset necrotizing encephalopathy with lethal outcome. J Inherit Metab Dis 28: 229-233.
- Dirik E,YisU, PasaogluG, Chambaz C, Baumgartner MR (2008) Recurrent attacks of status epilepticus as predominant symptom in 3-methylcrotonyl-CoA carboxylase deficiency. Brain Dev 30: 218-220.
- Steen C, Baumgartner ER, Duran M, Lehnert W, Suormala T, et al. (1999) Metabolic stroke in isolated 3-methylcrotonyl-CoA carboxylase deficiency. Eur J Pediatr 158: 730-733.
- Wiesmann UN, Suormala T, Pfenninger J, Baumgartner ER (1998) Partial 3-methylcrotonyl-CoA carboxylase deficiency in an infant with fatal outcome due to progressive respiratory failure. Eur J Pediatr 157: 225-229.
- Leon-Del-Rio A, Gravel RA (1994) Sequence requirements for the biotinylation of carboxyl-terminal fragments of human propionyl-CoA carboxylase alpha subunit expressed in Escherichia coli. J BiolChem 269: 22964-22968.
- Murin R, Verleysdonk S, Rapp M, Hamprecht B (2006)Immunocytochemical localization of 3-methylcrotonyl-CoA carboxylase in cultured ependymal, microglial and oligodendroglial cells. J Neurochem 97: 1393-1402.
- Rodriguez-Melendez R, Cano S, Mendez ST, Velazquez A (2001) Biotin regulates the genetic expression of holocarboxylasesynthetase and mitochondrial carboxylases in rats. J Nutr 131: 1909-1913.
- Stadler SC, Polanetz R, Meier S, Mayerhofer PU, Herrmann JM, et al. (2005) Mitochondrial targeting signals and mature peptides of 3-methylcrotonyl-CoA carboxylase. BiochemBiophys Res Commun 334: 939-946.
- Baumgartner MR, Almashanu S, Suormala T, Obie C, Cole RN, et al. (2001) The molecular basis of human 3-methylcrotonyl-CoA carboxylase deficiency. J Clin Invest 107: 495-504.
- Gallardo ME, Desviat LR, Rodriguez JM, Esparza-Gordillo J, Perez-Cerda C, et al. (2001) The molecular basis of 3-methylcrotonylglycinuria, a disorder of leucine catabolism. Am J Hum Genet 68: 334-346.
- Holzinger A, Röschinger W, Lagler F, Mayerhofer PU, Lichtner P, et al. (2001) Cloning of the human MCCA and MCCB genes and mutations therein reveal the molecular cause of 3-methylcrotonyl-CoA: carboxylase deficiency. Hum Mol Genet 10: 1299-1306.
- Shepard PJ, Barshop BA, Baumgartner MR, Hansen JB, Jepsen K, et al (2014) Consanguinity and rare mutations outside of MCCC genes underlie nonspecific phenotypes of MCCD. Genet Med 17: 660-667.
- Wu AHB (2006)Tietz Clinical guide to laboratory tests (4thedn.) Saunders Elsevier.
- Hoffman GF,Feyh P (2008) Organic Acid Analysis.In: Blau N, Duran M and Gibson KM (eds.) Laboratory Guide to the Methods in Biochemical Genetics. Springer Berlin Heidelberg, pp: 27-44.
- Rinaldo P (2008) Organic Acids.In: Blau N, Duran M and Gibson KM (eds.) Laboratory Guide to the Methods in Biochemical Genetics. Springer Berlin Heidelberg, pp: 137-169.
- Millington DS (2008) Tandem mass spectrometry in clinical diagnosis.In: Blau N, Duran M and Gibson KM (eds.) Laboratory Guide to the Methods in Biochemical Genetics. Springer Berlin Heidelberg, pp: 57-75.
- Galjaard H (1980) Genetic metabolic diseases: Early diagnosis and prenatal analysis, Elsevier/North-Holland biomedical press, Amsterdam.
- Suormala T, Wick H, Bonjour JP, Baumgartner ER (1985) Rapid differential diagnosis of carboxylase deficiencies and evaluation for biotin-responsiveness in a single blood sample. ClinicaChimicaActa 145: 151-162.
- Suormala T, Fowler B, Jakobs C, Duran M, Lehnert A (1997)Fivepatients with a biotin-responsive defect in holocarboxylase formation: evaluation of responsiveness to biotin therapy in vivo and comparative biochemical studies in vitro. Pediatr Res 41: 666-673.
- Suormala T, Fowler B, Jakobs C, Duran M, Lehnert W, et al.(1998) Late-onset holocarboxylasesynthetase deficiency: pre- and post-natal diagnosis and evaluation of effectiveness of antenatal biotin therapy. Eur J Pediatr 157: 570-575.
- Suormala T, Fowler B, Jakobs C, Duran M, Lehnert A(1997)Five patients with a biotin-responsive defect in holocarboxylase formation: evaluation of responsiveness to biotin therapy in vivo and comparative biochemical studies in vitro. Pediatr Res 41: 666-673.
- Wolf B,Feldman GL (1982) The biotin-dependent carboxylase deficiencies. Am J Hum Genet 34: 699-716.
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947-2948.
- Affymetrix(2014) Annotation files.
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, AntonellisKJ, et al.(2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249-264.
- Livak KJ, Flood SJ, Marmaro J, Giusti W, Deetz K (1995) Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl 4: 357-362.
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta DeltaC(T)) Method. Methods 25: 402-408.
- Mueller P, Schulze A, Schindler I, Ethofer T, Buehrdel P, et al. (2003) Validation of an ESI-MS/MS screening method for acylcarnitine profiling in urine specimens of neonates, children, adolescents and adults. ClinicaChimicaActa 327: 47-57.
- Sharif O, Knapp S (2008) From expression to signaling: roles of TREM-1 and TREM-2 in innate immunity and bacterial infection. Immunobiology 213: 701-713.
- Bouchon A, Hernández-Munain C, Cella M, Colonna M (2001) A DAP12-mediated Pathway Regulates expression of CC Chemokine Receptor 7 and Maturation of Human Dendritic Cells. J Exp Med 194: 1111-1122.
- Somlyo AP, Somlyo AV (1994) Signal transduction and regulation in smooth muscle. Nature 372: 231-236.
- Abisambra JF, Jinwal UK, Jones JR, Blair LJ, Koren J, et al. (2011) Exploiting the diversity of the heat-shock protein family for primary and secondary tauopathy therapeutics. CurrNeuropharmacol 9: 623-631.
- Moura AP, Ribeiro CA, Zanatta A, Busanello EN, Tonin AM, et al.(2012) 3-Methylcrotonylglycine disrupts mitochondrial energy homeostasis and inhibits synaptic Na(+),K (+)-ATPase activity in brain of young rats. Cell MolNeurobiol 32: 297-307.
- Wajner M, Latini A, Wyse AT, Dutra-Filho CS (2004) The role of oxidative damage in the neuropathology of organic acidurias: insights from animal studies. J Inherit Metab Dis 27: 427-448.
- ZanattA, Moura AP, Tonin AM, Knebel LA, Grings M, et al.(2013) Neurochemical evidence that the metabolites accumulating in 3-methylcrotonyl-CoA carboxylase deficiency induce oxidative damage in cerebral cortex of young rats. Cell MolNeurobiol 33: 137-146.
- Divito EB, Cascio M (2013) Metabolism, physiology, and analyses of primary fatty acid amides. Chem Rev 113: 7343-7353.
- Calder PC (2015) Marine omega-3 fatty acids and inflammatory processes: Effects, mechanisms and clinical relevance. BiochimBiophysActa 1851: 469-484.
- Ribeiro CA, Hickmann FH,Wajner M(2011) Neurochemical evidence that 3-methylglutaric acid inhibits synaptic Na+,K+-ATPase activity probably through oxidative damage in brain cortex of young rats. Int J DevNeurosci 29: 1-7.
- Botta A, Laher I, Beam J, Decoffe D, Brown K, et al. (2013) Short term exercise induces PGC-1alpha, ameliorates inflammation and increases mitochondrial membrane proteins but fails to increase respiratory enzymes in aging diabetic hearts. PLoS One 8: e70248.
- Sitta A, Ribas GS, Mescka CP, Barschak AG, Wajner M, et al. (2014) Neurological damage in MSUD: the role of oxidative stress. Cell MolNeurobiol 34: 157-165.
- Sirtori LR, Dutra-Filho CS, Fitarelli D, Sitta A, Haeser A, et al. (2005) Oxidative stress in patients with phenylketonuria. BiochimBiophysActa 1740: 68-73.
- Leipnitz G, Seminotti B, Amaral AU, de Bortoli G, Solano A, et al. (2008) Induction of oxidative stress by the metabolites accumulating in 3-methylglutaconic aciduria in cerebral cortex of young rats. Life Sci 82: 652-662.
- Wajner M, Goodman SI (2011) Disruption of mitochondrial homeostasis in organic acidurias: insights from human and animal studies. J BioenergBiomembr 43: 31-38.
- Gardner PR (2002) Aconitase: sensitive target and measure of superoxide. Methods Enzymol 349: 9-23.
- Murphy MP (2009) How mitochondria produce reactive oxygen species. Biochem J 417: 1-13.
- Babior BM (1999) NADPH oxidase: an update. Blood 93: 1464-1476.
- Reuter S1 Gupta SC, Chaturvedi MM, Aggarwal BB (2010) Oxidative stress, inflammation, and cancer: how are they linked? Free RadicBiol Med 49: 1603-1616.
- Gregersen N, Olsen RK (2010) Disease mechanisms and protein structures in fatty acid oxidation defects. J Inherit Metab Dis 33: 547-553.
- Johnson AR, Milner JJ,Makowski L (2012)The inflammation highway: metabolism accelerates inflammatory traffic in obesity. Immunol Rev 249: 218-238.
- Muñoz A, Costa M (2013) Nutritionally mediated oxidative stress and inflammation. Oxid Med Cell Longev 2013: 610950.
- Greene ER, Huang S, Serhan CN,Panigrahy D (2011) Regulation of inflammation in cancer by eicosanoids. Prostaglandins Other Lipid Mediat 96: 27-36.
- Di Filippo M, Chiasserini D, Tozzi A, Picconi B, Calabresi P (2010) Mitochondria and the link between neuroinflammation and neurodegeneration. J Alzheimers Dis 20: S369-S379.
- Filiou MD, Arefin AS, Moscato P,Graeber MB (2014) 'Neuroinflammation' differs categorically from inflammation: transcriptomes of Alzheimer's disease, Parkinson's disease, schizophrenia and inflammatory diseases compared. Neurogenetics 15: 201-212.
- Kones R(2010)Mitochondrial therapy for Parkinson's disease: neuroprotectivepharmaconutrition may be disease-modifying. ClinPharmacol 2: 185-198.
- Blass JP,Sheu RK,Gibson GE (2000) Inherent abnormalities in energy metabolism in Alzheimer disease. Interaction with cerebrovascular compromise. Ann N Y AcadSci 903: 204-221.
- Calingasan NY, Ho DJ, Wille EJ, Campagna MV, Ruan JD (2008) Influence of mitochondrial enzyme deficiency on adult neurogenesis in mouse models of neurodegenerative diseases. Neuroscience 153: 986-996.
- Farooqui AA, Ong WY,Horrocks LA (2006) Inhibitors of brain phospholipase A2 activity: their neuropharmacological effects and therapeutic importance for the treatment of neurologic disorders. Pharmacol Rev 58: 591-620.
- Sun GY, Shelat PB, Jensen MB, He Y, Sun AY, et al. (2010) Phospholipases A2 and inflammatory responses in the central nervous system. Neuromolecular Med 12: 133-148.
- Korotkova M, Jakobsson PJ (2014) Persisting eicosanoid pathways in rheumatic diseases. Nat Rev Rheumatol 10: 229-241.
- Quach ND, Arnold RD, Cummings BS (2014) Secretory phospholipase A2 enzymes as pharmacological targets for treatment of disease. BiochemPharmacol 90: 338-348.
- Kang L, Wang YS, Uppalapati SR, Wang K, Tang Y, et al. (2008) Overexpression of a fatty acid amide hydrolase compromises innate immunity in Arabidopsis. Plant J 56: 336-349.
- Bashashati M, Storr MA, Nikas SP, Wood JT, Godlewski G(2012) Inhibiting fatty acid amide hydrolase normalizes endotoxin-induced enhanced gastrointestinal motility in mice. Br J Pharmacol 165: 1556-1571.
- Naughton SS, Mathai ML, Hryciw DH, McAinch AJ (2013) Fatty Acid modulation of the endocannabinoid system and the effect on food intake and metabolism. Int J Endocrinol 2013: 361895.
- Kaczocha M, Lin Q, Nelson LD, McKinney MK, Cravatt BF, et al. (2012)Anandamide externally added to lipid vesicles containing trapped fatty acid amide hydrolase (FAAH) is readily hydrolyzed in a sterol-modulated fashion. ACS ChemNeurosci 3: 364-368.
- Dennis EA (2000) Phospholipase A2 in eicosanoid generation. Am J RespirCrit Care Med 161: S32-35.
- Murakami M, Taketomi Y, Sato H, Yamamoto K (2011) Secreted phospholipase A2 revisited. J Biochem 150: 233-255.
- Ryan VH, Primiani CT, Rao J, Ahn K, Rapoport SI, et al. (2014) Coordination of gene expression of arachidonic and docosahexaenoic acid cascade enzymes during human brain development and aging. PLoS One 9: e100858.
- Libby P, Tabas I, Fredman G, Fisher EA (2014) Inflammation and its resolution as determinants of acute coronary syndromes. Circ Res 114: 1867-1879.
- Serhan CN, Petasis NA (2011) Resolvins and protectins in inflammation resolution. Chem Rev 111: 5922-5943.
- Norris PC, Gosselin D, Reichart D, Glassb CK, Dennis EA (2014) Phospholipase A2 regulates eicosanoid class switching during inflammasome activation. PNAS 111: 12746-12751.
- Savari S, Vinnakota K, Zhang Y,Sjolander A(2014)Cysteinylleukotrienes and their receptors: bridging inflammation and colorectal cancer. World J Gastroenterol 20: 968-977.
- Brightling CE, Ward R, Woltmann G, Bradding P, Sheller JR, et al.(2000) Induced sputum inflammatory mediator concentrations in eosinophilic bronchitis and asthma. Am J RespirCrit Care Med 162: 878-882.
- Levy BD, Clish CB, Schmidt B, Gronert K,Serhan CN(2001) Lipid mediator class switching during acute inflammation: signals in resolution. Nature Immunol2: 612-619.
- Levy BD, Kohli P,Gotlinger K, Haworth O, Hong S, et al. (2007) Protectin D1 is generated in asthma and dampens airway inflammation and hyperresponsiveness. J Immunol 178: 496-502.
- Pavord ID, Ward R, Woltmann G, Wardlaw AJ, Sheller JR, et al. (1999) Induced sputum eicosanoid concentrations in asthma. AmJRespirCrit Care Med 160: 1905-1909.
- Glaser C, Lattka E, Rzehak P, Steer C, Koletzko B (2011) Genetic variation in polyunsaturated fatty acid metabolism and its potential relevance for human development and health. Matern Child Nutr 7: 27-40.
- Lattka E, Illig T, Heinrich J,Koletzko B(2009) FADS gene cluster polymorphisms: important modulators of fatty acid levels and their impact on atopic diseases. J NutrigenetNutrigenomics2: 119-128.
- Lindskov R, Hølmer G (1992) Polyunsaturated fatty acids in plasma, red blood cells and mononuclear cell phospholipids of patients with atopic dermatitis. Allergy 47: 517-521.
- Ribeiro CA, Balestro F, Grando V, Wajner M (2007) Isovaleric acid reduces Na+, K+-ATPase activity in synaptic membranes from cerebral cortex of young rats. Cell MolNeurobiol 27: 529-540.
- Wortmann SB, Kluijtmans LA, Engelke UF, Wevers RA, Morava E (2012) The 3-methylglutaconic acidurias: what's new? J Inherit Metab Dis 35: 13-22.
- Cherlyn SY, Woon PS, Liu JJ, Ong WY, Tsai GC(2010) Genetic association studies of glutamate, GABA and related genes in schizophrenia and bipolar disorder: a decade of advance. NeurosciBiobehav Rev 34: 958-977.
- Zandberg L, van Dyk HC, van der Westhuizen FH, van Dijk AA (2016)A 3-methylcrotonyl-CoA carboxylase deficient human skin fibroblast transcriptome reveals underlying mitochondrial dysfunction and oxidative stress. Int J Biochem Cell Biol 78: 116-129.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences